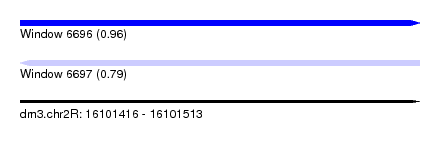
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,101,416 – 16,101,513 |
| Length | 97 |
| Max. P | 0.955915 |

| Location | 16,101,416 – 16,101,513 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.25 |
| Shannon entropy | 0.57837 |
| G+C content | 0.54924 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
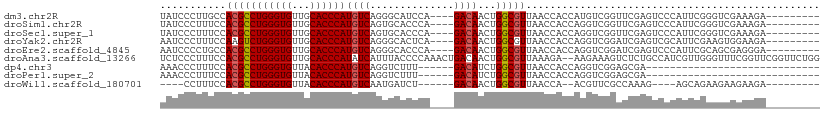
>dm3.chr2R 16101416 97 + 21146708 ---------UCUUUCGACCCGAAUGGGACUCGAACCGACAUGGUGGUUAACGCCAGUUGUC----UGGAUGCCCUGACAUGGGUGCAACACCCAGGCGUGGCAAGGGAUA ---------...((((((((....)))..)))))(((((((((((.....)))))..))))----....((((.((...((((((...))))))..)).)))).)).... ( -36.20, z-score = -1.21, R) >droSim1.chr2R 14760269 97 + 19596830 ---------UCUUUCGACCCGAAUGGGACUCGAACCGACCUGGUGGUUAACGCCAGUUGUC----UGGGUGCACUGACAUGGGUGCAACACCCAGGCGUGGAAAGGGAUA ---------...((((((((....)))..)))))((...((((((.....))))))..(((----((((((((((......)))))...)))))))).......)).... ( -38.30, z-score = -1.73, R) >droSec1.super_1 13662011 97 + 14215200 ---------UCUUUCGACCCGAAUGGGACUCGAACCGACCUGGUGGUUAACGCCAGUUGUC----UGGGUGCACUGACAUGGGUGCAACACCCAGGCGUGGAAAGGGAUA ---------...((((((((....)))..)))))((...((((((.....))))))..(((----((((((((((......)))))...)))))))).......)).... ( -38.30, z-score = -1.73, R) >droYak2.chr2R 8036522 97 - 21139217 ---------UCUUCCACUUCGAAUGCGACUCGAUCCGACCUGGUGGUUAACGCCAGUUGUC----UGAGUGCCCUGACAUGGGUGCAACACCCAGACUUGGAAAGGGAUU ---------..((((...((((.......))))(((((.((((((.....))))))..(((----((.(..(((......)))..)......))))))))))..)))).. ( -32.70, z-score = -1.46, R) >droEre2.scaffold_4845 10251745 97 + 22589142 ---------UCCCUCGCUGCGAAUGGGACUCGAUCCGACCUGGUGGUUAACGCCAGUUGUC----UGGGUGCCCUGACAUGGGUGCAACACCCAGGCGUGGCAGGGGAUU ---------((((..((..((..((((.......(((((((((((.....))))))..)))----.))(..(((......)))..)....))))..))..))..)))).. ( -49.70, z-score = -3.42, R) >droAna3.scaffold_13266 10644299 108 - 19884421 CCAGAACCGAACCGAAACCCAACGAUGGCAGAGACUUUCUU--UCUUUAACGCCAGUUGUCAGUUUGGGGUAAAUGAUAUGGGUGCAACACCCAGGCGUGGAAAGGGAGA ..................(((....)))......((..(((--(((...(((((...(((((.((......)).)))))((((((...)))))))))))))))))..)). ( -30.50, z-score = -0.40, R) >dp4.chr3 13238177 75 + 19779522 -----------------------------UCGCUCCGACCUGGUGGUUAACGCCAGAUGUC------AAAGACCUGACAUGGGUGUAACACCCAGGCGUGGAAAGGGUUU -----------------------------((..((((.(((((..((((.((((..(((((------(......)))))).))))))))..)))))..))))..)).... ( -29.60, z-score = -2.29, R) >droPer1.super_2 650588 75 - 9036312 -----------------------------UCGCUCCGACCUGGUGGUUAACGCCAGAUGUC------AAAGACCUGACAUGGGUGUAACACCCAGGCGUGGAAAGGGUUU -----------------------------((..((((.(((((..((((.((((..(((((------(......)))))).))))))))..)))))..))))..)).... ( -29.60, z-score = -2.29, R) >droWil1.scaffold_180701 2588454 85 + 3904529 ---------UCUUCUUCUUCUGCU----CUUUGGCGAACGU--UGGUUAACGCCAGUUGUC------AGAUCAUUGACAUGGGUGUAACACCCAGGCGUGGAAAGG---- ---------........(((..(.----..(..((....))--..).....(((...((((------((....))))))((((((...))))))))))..)))...---- ( -24.90, z-score = -1.00, R) >consensus _________UCUUUCGACCCGAAUGGGACUCGAACCGACCUGGUGGUUAACGCCAGUUGUC____UGGGUGCCCUGACAUGGGUGCAACACCCAGGCGUGGAAAGGGAUA ..................................(((((......))).(((((...((((..............))))((((((...))))))))))).....)).... (-16.86 = -17.26 + 0.40)

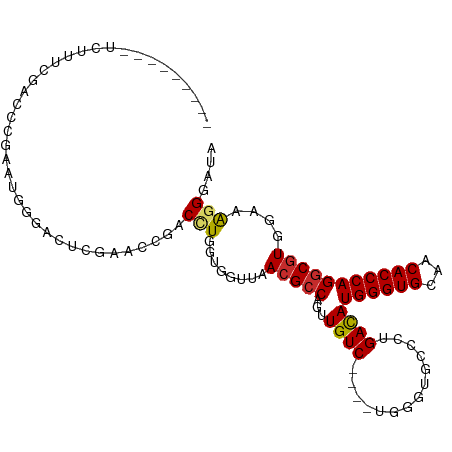
| Location | 16,101,416 – 16,101,513 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 70.25 |
| Shannon entropy | 0.57837 |
| G+C content | 0.54924 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.41 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16101416 97 - 21146708 UAUCCCUUGCCACGCCUGGGUGUUGCACCCAUGUCAGGGCAUCCA----GACAACUGGCGUUAACCACCAUGUCGGUUCGAGUCCCAUUCGGGUCGAAAGA--------- ...(((.((((..((.((((((...)))))).))...))))....----((((((((((((........)))))))))...)))......)))((....))--------- ( -31.60, z-score = -0.59, R) >droSim1.chr2R 14760269 97 - 19596830 UAUCCCUUUCCACGCCUGGGUGUUGCACCCAUGUCAGUGCACCCA----GACAACUGGCGUUAACCACCAGGUCGGUUCGAGUCCCAUUCGGGUCGAAAGA--------- .....(((((...((((((..(((((((........))))(((((----(....)))).)).)))..)))))).((..(((((...)))))..))))))).--------- ( -32.50, z-score = -1.37, R) >droSec1.super_1 13662011 97 - 14215200 UAUCCCUUUCCACGCCUGGGUGUUGCACCCAUGUCAGUGCACCCA----GACAACUGGCGUUAACCACCAGGUCGGUUCGAGUCCCAUUCGGGUCGAAAGA--------- .....(((((...((((((..(((((((........))))(((((----(....)))).)).)))..)))))).((..(((((...)))))..))))))).--------- ( -32.50, z-score = -1.37, R) >droYak2.chr2R 8036522 97 + 21139217 AAUCCCUUUCCAAGUCUGGGUGUUGCACCCAUGUCAGGGCACUCA----GACAACUGGCGUUAACCACCAGGUCGGAUCGAGUCGCAUUCGAAGUGGAAGA--------- ..(((((.(((..(((((((((.....(((......)))))))))----)))..((((.(.....).))))...)))((((((...)))))))).)))...--------- ( -32.60, z-score = -1.22, R) >droEre2.scaffold_4845 10251745 97 - 22589142 AAUCCCCUGCCACGCCUGGGUGUUGCACCCAUGUCAGGGCACCCA----GACAACUGGCGUUAACCACCAGGUCGGAUCGAGUCCCAUUCGCAGCGAGGGA--------- ..((((((((...((((((..((((.(((((((((.((....)).----))))..))).))))))..)))))).((((...)))).....))))...))))--------- ( -35.60, z-score = -0.66, R) >droAna3.scaffold_13266 10644299 108 + 19884421 UCUCCCUUUCCACGCCUGGGUGUUGCACCCAUAUCAUUUACCCCAAACUGACAACUGGCGUUAAAGA--AAGAAAGUCUCUGCCAUCGUUGGGUUUCGGUUCGGUUCUGG .........(((.(((((((((...))))))..............((((((...(..(((....(((--.........))).....)))..)...)))))).)))..))) ( -24.80, z-score = 0.54, R) >dp4.chr3 13238177 75 - 19779522 AAACCCUUUCCACGCCUGGGUGUUACACCCAUGUCAGGUCUUU------GACAUCUGGCGUUAACCACCAGGUCGGAGCGA----------------------------- .....(.((((..((((((..((((.(((((((((((....))------))))..))).))))))..)))))).)))).).----------------------------- ( -25.10, z-score = -1.56, R) >droPer1.super_2 650588 75 + 9036312 AAACCCUUUCCACGCCUGGGUGUUACACCCAUGUCAGGUCUUU------GACAUCUGGCGUUAACCACCAGGUCGGAGCGA----------------------------- .....(.((((..((((((..((((.(((((((((((....))------))))..))).))))))..)))))).)))).).----------------------------- ( -25.10, z-score = -1.56, R) >droWil1.scaffold_180701 2588454 85 - 3904529 ----CCUUUCCACGCCUGGGUGUUACACCCAUGUCAAUGAUCU------GACAACUGGCGUUAACCA--ACGUUCGCCAAAG----AGCAGAAGAAGAAGA--------- ----.((((..(((((((((((...))))))(((((......)------))))...)))))......--..((((......)----))).)))).......--------- ( -25.10, z-score = -2.33, R) >consensus AAUCCCUUUCCACGCCUGGGUGUUGCACCCAUGUCAGGGCACCCA____GACAACUGGCGUUAACCACCAGGUCGGAUCGAGUCCCAUUCGGGUCGAAAGA_________ ...........(((((((((((...))))))((((..............))))...)))))................................................. (-15.28 = -15.41 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:38 2011