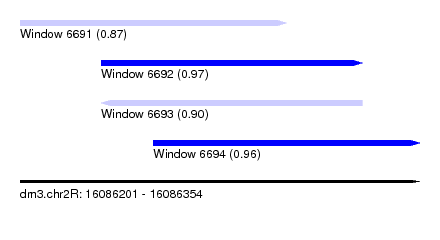
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,086,201 – 16,086,354 |
| Length | 153 |
| Max. P | 0.971352 |

| Location | 16,086,201 – 16,086,303 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Shannon entropy | 0.14943 |
| G+C content | 0.51905 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -30.64 |
| Energy contribution | -31.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
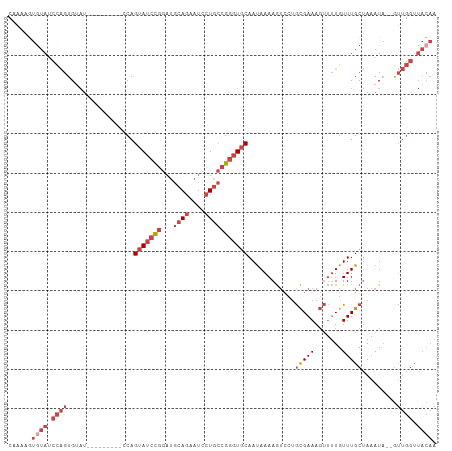
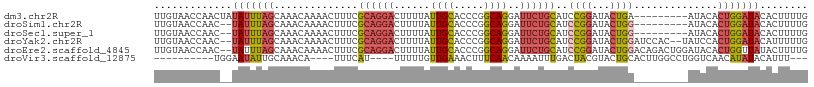
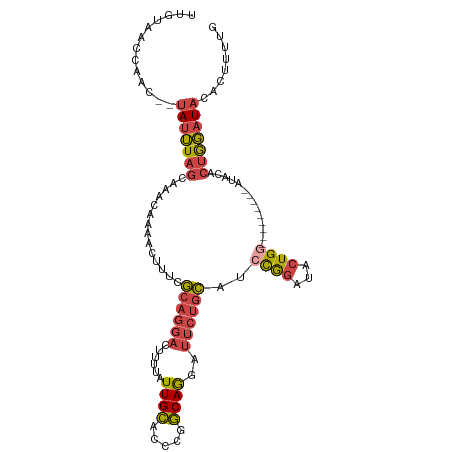
>dm3.chr2R 16086201 102 + 21146708 GGAAUGCUCGCUGACUUGGCAAGCUGGAGGGCAAAAGUGUAUCCAGUGUAU---------UCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG .......((((.(((((.....((((((..(((....))).))))))....---------...(((((((...((((....))))))))))).....)))))..))))... ( -35.40, z-score = -2.14, R) >droSim1.chr2R 14743933 102 + 19596830 GCCAUGCUCGCUGACUUGGCAAGCUGAAGGGCAAAAGUGUAUCCAGUGUAU---------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG ((((.(((.(((.....))).))))).(((((.....(((((((.....((---------((.......))))((((....)))).)))))))......)))))))..... ( -31.60, z-score = -1.11, R) >droSec1.super_1 13647008 102 + 14215200 GCCAUGCUCGCUGACUUGGCAAGCUGGAGGGCAAAAGUGUAUCCAGUGUAU---------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG .......((((.(((((((...((((((..(((....))).))))))....---------)).(((((((...((((....))))))))))).....)))))..))))... ( -37.60, z-score = -2.48, R) >droYak2.chr2R 8020651 109 - 21139217 GGCAUGCUCGCUGACUUGGCAAGCUGGAGGGCAAAAAUGUAUCCAGUGGAUA--GUGGAUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG .......((((.(((((.(((..((((((((......(((((((...(((((--.((....)).))))))))))))...)))).)))).))).....)))))..))))... ( -41.50, z-score = -2.57, R) >droEre2.scaffold_4845 10236468 111 + 22589142 GGCAUGCUCGCUGACUUGGCAAUCUGGAGGGCAAAAGUAUAACCAGUGUAUCCAGUCUGUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG .......((((.(((((((((..(((((..(((...(.....)...))).)))))..))))..(((((((...((((....))))))))))).....)))))..))))... ( -38.30, z-score = -2.17, R) >consensus GGCAUGCUCGCUGACUUGGCAAGCUGGAGGGCAAAAGUGUAUCCAGUGUAU_________CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAG .......((((.(((((.....((((((..(((....))).))))))................(((((((...((((....))))))))))).....)))))..))))... (-30.64 = -31.44 + 0.80)

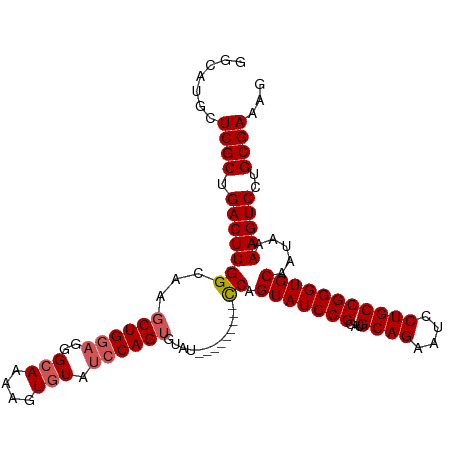
| Location | 16,086,232 – 16,086,332 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.50093 |
| G+C content | 0.39909 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -13.38 |
| Energy contribution | -15.80 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
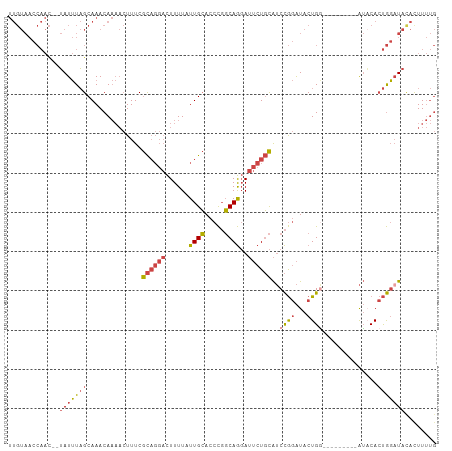
>dm3.chr2R 16086232 100 + 21146708 CAAAAGUGUAUCCAGUGUAU---------UCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUAUAGUUGGUUACAA ......((((.(((((((((---------(..(((((((...((((....)))))))))))............(((((.......)))))..)))))).)))).)))). ( -26.60, z-score = -2.58, R) >droSim1.chr2R 14743964 98 + 19596830 CAAAAGUGUAUCCAGUGUAU---------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAA ......((((.(((((((..---------...(((((((...((((....)))))))))))............(((((.......)))))...)))--.)))).)))). ( -23.90, z-score = -1.68, R) >droSec1.super_1 13647039 98 + 14215200 CAAAAGUGUAUCCAGUGUAU---------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAA ......((((.(((((((..---------...(((((((...((((....)))))))))))............(((((.......)))))...)))--.)))).)))). ( -23.90, z-score = -1.68, R) >droYak2.chr2R 8020682 105 - 21139217 CAAAAAUGUAUCCAGUGGAUA--GUGGAUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAA ......((((.((((((..((--(..(((...(((((((...((((....)))))))))))............((....))...)))..)))..))--.)))).)))). ( -29.70, z-score = -2.31, R) >droEre2.scaffold_4845 10236499 107 + 22589142 CAAAAGUAUAACCAGUGUAUCCAGUCUGUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAA ...............((((.((((.((((...(((((((...((((....)))))))))))............(((((.......)))))...)))--))))).)))). ( -26.50, z-score = -1.93, R) >droVir3.scaffold_12875 17204984 88 - 20611582 ---AAAUGUAUAUGUUGACCAGGCCAAGUGCAGUACGUAGUCAAAUUUUGUUGAAAGUUUCAACAAAAA----AUGAAA----UGUUUGCAAUAUUCCA---------- ---..................((.....(((((.((((..(((..(((((((((.....))))))))).----.))).)----)))))))).....)).---------- ( -19.50, z-score = -2.20, R) >consensus CAAAAGUGUAUCCAGUGUAU_________CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA__GUUGGUUACAA ......((((.((((.................(((((((...((((....)))))))))))............(((((.......))))).........)))).)))). (-13.38 = -15.80 + 2.42)

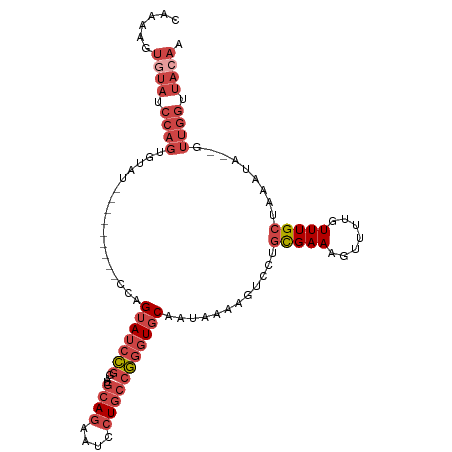
| Location | 16,086,232 – 16,086,332 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.50093 |
| G+C content | 0.39909 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -8.94 |
| Energy contribution | -10.37 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16086232 100 - 21146708 UUGUAACCAACUAUAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGA---------AUACACUGGAUACACUUUUG .((((.(((....(((((((..............((((........))))..(((((((....))))...)))...))))---------)))...))).))))...... ( -20.50, z-score = -2.29, R) >droSim1.chr2R 14743964 98 - 19596830 UUGUAACCAAC--UAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGG---------AUACACUGGAUACACUUUUG .((((.(((..--.....................((((((......((((.....))))..))))))((((((...))))---------))....))).))))...... ( -20.70, z-score = -2.02, R) >droSec1.super_1 13647039 98 - 14215200 UUGUAACCAAC--UAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGG---------AUACACUGGAUACACUUUUG .((((.(((..--.....................((((((......((((.....))))..))))))((((((...))))---------))....))).))))...... ( -20.70, z-score = -2.02, R) >droYak2.chr2R 8020682 105 + 21139217 UUGUAACCAAC--UAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGGAUCCAC--UAUCCACUGGAUACAUUUUUG .((((.(((..--.....................((((((......((((.....))))..))))))....(((((.((....)).--)))))..))).))))...... ( -23.50, z-score = -2.23, R) >droEre2.scaffold_4845 10236499 107 - 22589142 UUGUAACCAAC--UAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGGACAGACUGGAUACACUGGUUAUACUUUUG .((((((((..--((((((((.............((((((......((((.....))))..)))))).(((((...)))))..).)))))))...))))))))...... ( -26.40, z-score = -2.70, R) >droVir3.scaffold_12875 17204984 88 + 20611582 ----------UGGAAUAUUGCAAACA----UUUCAU----UUUUUGUUGAAACUUUCAACAAAAUUUGACUACGUACUGCACUUGGCCUGGUCAACAUAUACAUUU--- ----------.((.....((((.((.----..(((.----.(((((((((.....)))))))))..)))....))..)))).....))..................--- ( -16.10, z-score = -1.88, R) >consensus UUGUAACCAAC__UAUUUAGCAAACAAAACUUUCGCAGGACUUUUAUUGCACCCGGCAGGAUUCUGCAUCCGGAUACUGG_________AUACACUGGAUACACUUUUG .((((.(((.........................((((((......((((.....))))..))))))..((((...))))...............))).))))...... ( -8.94 = -10.37 + 1.42)
| Location | 16,086,252 – 16,086,354 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Shannon entropy | 0.11973 |
| G+C content | 0.41579 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16086252 102 + 21146708 -------UCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUAUAGUUGGUUACAAAUUUUGUAAACUCCAGGCUUUU -------...(((((((...((((....)))))))))))...(((((.(((((....)....((((((.((((....(((....))))))).))))))..))))))))) ( -28.10, z-score = -2.26, R) >droSim1.chr2R 14743984 99 + 19596830 -------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAAAUUUUGUAAACUCCAGGCUUU- -------...(((((((...((((....)))))))))))....((((.(((((....)....((((((.(((..--.(((....)))..)))))))))..))))))))- ( -28.30, z-score = -2.30, R) >droSec1.super_1 13647059 99 + 14215200 -------CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAAAUUUUGUAAACUCCAGGCUUU- -------...(((((((...((((....)))))))))))....((((.(((((....)....((((((.(((..--.(((....)))..)))))))))..))))))))- ( -28.30, z-score = -2.30, R) >droYak2.chr2R 8020702 106 - 21139217 AGUGGAUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAAAUUUUGUAAACUCCAGGCUUU- ..........(((((((...((((....)))))))))))....((((.(((((....)....((((((.(((..--.(((....)))..)))))))))..))))))))- ( -28.30, z-score = -1.42, R) >droEre2.scaffold_4845 10236521 106 + 22589142 AGUCUGUCCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA--GUUGGUUACAAAUGUUGUAAGCUCCAGGCUUC- ((((((....(((((((...((((....)))))))))))......(((..(((((....(((((..(((((...--.))))).)))))..))))).))).))))))..- ( -30.00, z-score = -2.11, R) >consensus _______CCAGUAUCCGGAUGCAGAAUCCUGCCGGGUGCAAUAAAAGUCCUGCGAAAGUUUUGUUUGCUAAAUA__GUUGGUUACAAAUUUUGUAAACUCCAGGCUUU_ ..........(((((((...((((....)))))))))))....((((.(((((....)....((((((.((((....(((....))))))).))))))..)))))))). (-26.48 = -26.72 + 0.24)

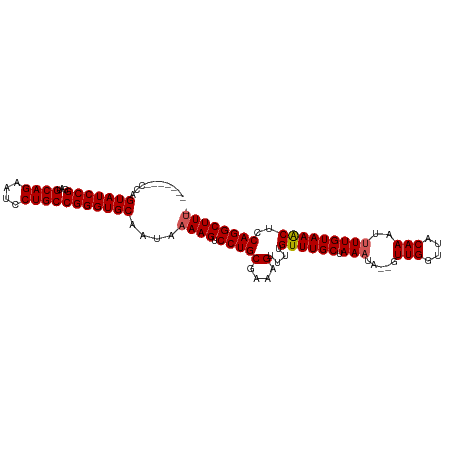
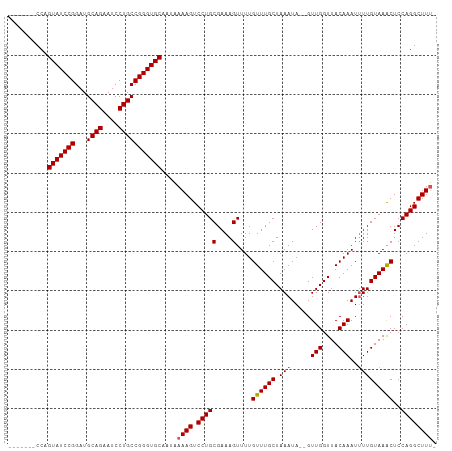
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:35 2011