
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,082,584 – 16,082,694 |
| Length | 110 |
| Max. P | 0.855627 |

| Location | 16,082,584 – 16,082,694 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Shannon entropy | 0.32716 |
| G+C content | 0.42636 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16082584 110 + 21146708 -AUUCCGUCC--CUCAGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAAUUCACGCAGUCG-- -.........--....((((((.-----...((..(((((....)))))))(((((((.(.....).)))))))......................))))))................-- ( -18.00, z-score = -1.63, R) >droSim1.chr2R 14740304 110 + 19596830 -AUUCCGUCC--CUCAGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUCACGCAGUCU-- -.........--....((((((.-----...((..(((((....)))))))(((((((.(.....).)))))))......................))))))................-- ( -18.00, z-score = -1.75, R) >droSec1.super_1 13643344 110 + 14215200 -AUUCCGUCC--CUCAGUCCGUU-----ACCGCCUGACAGUCUGGUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACUCAUUUCACGCAGUCG-- -....(((..--...(((((((.-----..((((.(.....).))))....(((((((.(.....).)))))))......................)))))))......)))......-- ( -16.80, z-score = -0.75, R) >droYak2.chr2R 8016980 115 - 21139217 -AUUCCGUCC--CUCAGUCCGUUCCGUUACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUCACGCAGUCU-- -.........--....((((((.........((..(((((....)))))))(((((((.(.....).)))))))......................))))))................-- ( -18.00, z-score = -1.51, R) >droEre2.scaffold_4845 10232960 110 + 22589142 -AUUCCGUCC--CUCAGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUAACGCAGUCU-- -.........--....((((((.-----...((..(((((....)))))))(((((((.(.....).)))))))......................))))))................-- ( -18.00, z-score = -1.72, R) >droAna3.scaffold_13266 10625685 110 - 19884421 ---CCUGUCC--GUGUGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUCACACAGUUCGU ---..(((((--((((((.....-----...((..(((((....)))))))(((((((.(.....).)))))))..................)))))))))))................. ( -26.80, z-score = -3.54, R) >dp4.chr3 13219424 109 + 19779522 -CGCUUGUAC--GCCUGUCCGUU-----CCCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUUACACAGCC--- -.(((((((.--...(((((((.-----...((..(((((....)))))))(((((((.(.....).)))))))......................))))))).....)))).))).--- ( -23.20, z-score = -3.17, R) >droPer1.super_2 631586 109 - 9036312 -CGCUUGUAC--GCCUGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUUACACAGCC--- -.(((((((.--...(((((((.-----...((..(((((....)))))))(((((((.(.....).)))))))......................))))))).....)))).))).--- ( -23.20, z-score = -3.03, R) >droWil1.scaffold_180701 2551797 109 + 3904529 -UUCUUGUACGAUUCCGUCCGUU-----ACCGCGUAACAGUCUGCUGCUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUACUCAUUUAUUUUAAACACGGGCCACAGCCACGCCG----- -...............((((((.-----.....(((.(((....))).)))(((((((.(.....).)))))))......................)))))).............----- ( -15.40, z-score = 0.67, R) >droVir3.scaffold_12875 17200869 97 - 20611582 -UUUGCGUCCG-GAGUGUCCGUU-----ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACAC---------------- -.....(((((-..((((.....-----...((..(((((....)))))))(((((((.(.....).)))))))..................)))).)))))..---------------- ( -21.50, z-score = -1.96, R) >droGri2.scaffold_15112 2734617 111 + 5172618 -CGCCCGUGCA-GAGUGUCCGUU-----ACCGCCUGACAGUCUGCUGCUGCAUAUUAAUGACUUUCUUUAAUGCCUUUUCCUCAUUUAUUUUACACACGGACACUUUUUCAACACUCG-- -.....(((.(-((((((((((.-----...((..(.(((....)))).)).((((((.(.....).)))))).......................))))))))))).....)))...-- ( -22.60, z-score = -2.34, R) >droMoj3.scaffold_6496 21714823 104 + 26866924 AUCUGCGUCCG-CUGUGUCCGUU-----ACCGCCUGACAGUCUGCUGCUGCAUAUUAAUGACUUUCUUUAAUGCCUUUUCCUCAUUUAUUUUACACACGGACACUUUUCA---------- ......(((((-.(((((.....-----...((..(.(((....)))).)).((((((.(.....).))))))...................))))))))))........---------- ( -19.90, z-score = -2.24, R) >consensus _AUUCCGUCC__CUCUGUCCGUU_____ACCGCCUGACAGUCUGCUGUUGCAUAUUAAUGACUUUCUUUAAUGUCUUUUCCUCAUUUAUUUUACACACGGACACAUUUCACGCAGUC___ ................((((((.........((....(((....)))..))(((((((.(.....).)))))))......................)))))).................. (-13.71 = -13.88 + 0.17)

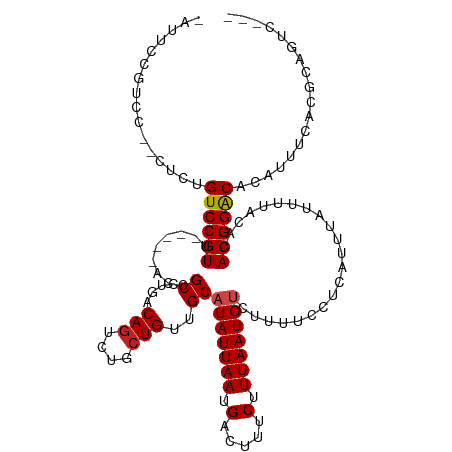
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:32 2011