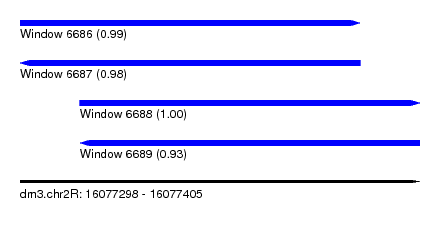
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,077,298 – 16,077,405 |
| Length | 107 |
| Max. P | 0.999398 |

| Location | 16,077,298 – 16,077,389 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Shannon entropy | 0.30802 |
| G+C content | 0.47230 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16077298 91 + 21146708 ------UUCUCCUGGUUCAAUC-CC--CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ------.......((......)-).--...........(((((((.....((((.....((((((((.....))))))))....)))).....))))))) ( -24.70, z-score = -2.48, R) >droSim1.chr2R 14735093 91 + 19596830 ------UUCUCCUGGCUCAAUC-CC--CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ------................-..--...........(((((((.....((((.....((((((((.....))))))))....)))).....))))))) ( -24.50, z-score = -2.10, R) >droSec1.super_1 13638084 91 + 14215200 ------UUCUUCUGGCUCAAUC-CC--CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ------................-..--...........(((((((.....((((.....((((((((.....))))))))....)))).....))))))) ( -24.50, z-score = -2.11, R) >droYak2.chr2R 8011305 91 - 21139217 ------UUCUUCUGGCUGAAUC-CC--CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ------........(((((((.-..--..........(((((......)))))......((((((((.....)))))))))))))))............. ( -28.50, z-score = -3.19, R) >droEre2.scaffold_4845 10227866 91 + 22589142 ------UUCUCCUGACUGCAUC-CC--CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ------.........(((((..-..--.........)))))((((.....((((.....((((((((.....))))))))....)))).....))))... ( -26.12, z-score = -2.87, R) >droAna3.scaffold_13266 10620944 98 - 19884421 UUGUUCUUGGCCAAGUCCAGUCGCC--UUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ...........(((((.(.(.((((--(...........))))).)....((((.....((((((((.....))))))))....)))).....).))))) ( -29.30, z-score = -2.01, R) >droMoj3.scaffold_6496 21709144 97 + 26866924 ---UUUUUGCUGCACUCUUGUUGUUGGCGUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCUGCAUUAUUUAUGCGGCUGCGCAGCGAUAACGCGCUUG ---...(((((((......((.(((((((((.........)))))))))..))......((((((((.....))))))))..)))))))........... ( -33.30, z-score = -2.02, R) >droWil1.scaffold_180701 2543491 75 + 3904529 -----------------------UC--UUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCACAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG -----------------------..--...........(((((((.....((((.....((((.(((.....))).))))....)))).....))))))) ( -19.10, z-score = -1.52, R) >consensus ______UUCUCCUGGCUCAAUC_CC__CUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUG ......................................(((((((.....((((.....((((((((.....))))))))....)))).....))))))) (-23.48 = -23.50 + 0.02)

| Location | 16,077,298 – 16,077,389 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Shannon entropy | 0.30802 |
| G+C content | 0.47230 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16077298 91 - 21146708 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG--GG-GAUUGAACCAGGAGAA------ ..((.(((((((((((....((((((((.....)))))))).....))).)).))))))))...........(--(.-.......)).......------ ( -26.00, z-score = -2.28, R) >droSim1.chr2R 14735093 91 - 19596830 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG--GG-GAUUGAGCCAGGAGAA------ ..((.(((((((((((....((((((((.....)))))))).....))).)).))))))))...........(--(.-.......)).......------ ( -25.00, z-score = -1.40, R) >droSec1.super_1 13638084 91 - 14215200 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG--GG-GAUUGAGCCAGAAGAA------ ..((.(((((((((((....((((((((.....)))))))).....))).)).))))))))...........(--(.-.......)).......------ ( -25.00, z-score = -1.65, R) >droYak2.chr2R 8011305 91 + 21139217 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG--GG-GAUUCAGCCAGAAGAA------ ...(((((....))).....((((((((.....))))))))......)).((((((..(((............--))-)..)))))).......------ ( -29.10, z-score = -3.13, R) >droEre2.scaffold_4845 10227866 91 - 22589142 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG--GG-GAUGCAGUCAGGAGAA------ .....(((((..((((....((((((((.....)))))))).....)))((((....))))...........)--..-)))))...........------ ( -25.50, z-score = -1.55, R) >droAna3.scaffold_13266 10620944 98 + 19884421 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAA--GGCGACUGGACUUGGCCAAGAACAA ...(((((....))).....((((((((.....))))))))......)).(((..((((((...........)--)))).......)..)))........ ( -29.01, z-score = -2.02, R) >droMoj3.scaffold_6496 21709144 97 - 26866924 CAAGCGCGUUAUCGCUGCGCAGCCGCAUAAAUAAUGCAGCUUAAAGAGCAGGUUGACGCCUACAAAAACAACGCCAACAACAAGAGUGCAGCAAAAA--- .............((((((((((.((((.....)))).)))..........((((.((.............)).)))).......))))))).....--- ( -23.92, z-score = -0.31, R) >droWil1.scaffold_180701 2543491 75 - 3904529 CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGUGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAA--GA----------------------- ..((.(((((((((((....((((((((.....)))))))).....))).)).))))))))............--..----------------------- ( -22.10, z-score = -2.68, R) >consensus CAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAG__GG_GAUUGAGCCAGGAGAA______ ..((.(((((((((((....((((((((.....)))))))).....))).)).))))))))....................................... (-23.03 = -23.05 + 0.02)
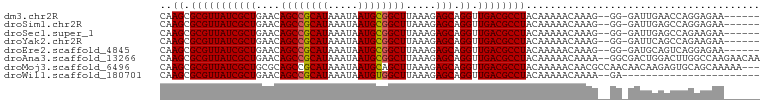

| Location | 16,077,314 – 16,077,405 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Shannon entropy | 0.29131 |
| G+C content | 0.47897 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16077314 91 + 21146708 ---CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACU------------- ---......((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))).........------------- ( -36.90, z-score = -5.38, R) >droSim1.chr2R 14735109 91 + 19596830 ---CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACU------------- ---......((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))).........------------- ( -36.90, z-score = -5.38, R) >droSec1.super_1 13638100 91 + 14215200 ---CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACU------------- ---......((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))).........------------- ( -36.90, z-score = -5.38, R) >droYak2.chr2R 8011321 91 - 21139217 ---CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACU------------- ---......((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))).........------------- ( -36.90, z-score = -5.38, R) >droEre2.scaffold_4845 10227882 91 + 22589142 ---CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACU------------- ---......((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))).........------------- ( -36.90, z-score = -5.38, R) >droAna3.scaffold_13266 10620964 99 - 19884421 UCGCCUUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACUCCAAC-------- (((.....(((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))----)))))...))).......-------- ( -37.40, z-score = -4.90, R) >droMoj3.scaffold_6496 21709166 108 + 26866924 ---GGCGUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCUGCAUUAUUUAUGCGGCUGCGCAGCGAUAACGCGCUUGCAAGAAAACGAGAAGAAAAAAAAGCCACGACUU ---(((....(((((((((((((.....((((.....((((((((.....))))))))....)))).....)))))))))))))..(.....)........)))....... ( -35.30, z-score = -1.53, R) >droGri2.scaffold_15112 2727956 103 + 5172618 ---GUCGUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUGCAGCGAUAACGCGCUUGCAAGCAAAAACGCCACGACUUCCACAAG----- ---((((((((((((((((((((...(((((......((((((((.....))))))))..)))).).....)))))))))....))))))..))))).........----- ( -37.80, z-score = -3.29, R) >droWil1.scaffold_180701 2543491 98 + 3904529 ---UCUUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCACAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAAAGCAAAGCGAAAAAAAAGGA---------- ---(((((((((((.((((((((.....((((.....((((.(((.....))).))))....)))).....))))))))))))))))).))..........---------- ( -30.60, z-score = -2.12, R) >droPer1.super_2 627131 98 - 9036312 -CAGGCCUUGUUUUUGGAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACUUUAAC-------- -..(((...(((((((.((((((.....((((.....((((((((.....))))))))....)))).....)))))).)))----)))))))...........-------- ( -38.10, z-score = -4.23, R) >dp4.chr3 13215005 98 + 19779522 -CAGGCCUUGUUUUUGGAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA----AAACGCCACGACUUGAAC-------- -..(((...(((((((.((((((.....((((.....((((((((.....))))))))....)))).....)))))).)))----)))))))...........-------- ( -38.10, z-score = -3.94, R) >consensus ___CCCUUUGUUUUUGUAGGCGUCAACCUGCUCUUUAAGCCGCAUUAUUUAUGCGGCUGUUCAGCGAUAACGCGCUUGCAA____AAACGCCACGACU_____________ ........(((((((((((((((.....((((.....((((((((.....))))))))....)))).....))))))))))....)))))..................... (-29.28 = -29.50 + 0.22)



| Location | 16,077,314 – 16,077,405 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Shannon entropy | 0.29131 |
| G+C content | 0.47897 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16077314 91 - 21146708 -------------AGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG--- -------------....(((((((((----((((.((((......))))....((((((((.....))))))))......)))))..)))))).))............--- ( -31.30, z-score = -2.16, R) >droSim1.chr2R 14735109 91 - 19596830 -------------AGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG--- -------------....(((((((((----((((.((((......))))....((((((((.....))))))))......)))))..)))))).))............--- ( -31.30, z-score = -2.16, R) >droSec1.super_1 13638100 91 - 14215200 -------------AGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG--- -------------....(((((((((----((((.((((......))))....((((((((.....))))))))......)))))..)))))).))............--- ( -31.30, z-score = -2.16, R) >droYak2.chr2R 8011321 91 + 21139217 -------------AGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG--- -------------....(((((((((----((((.((((......))))....((((((((.....))))))))......)))))..)))))).))............--- ( -31.30, z-score = -2.16, R) >droEre2.scaffold_4845 10227882 91 - 22589142 -------------AGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG--- -------------....(((((((((----((((.((((......))))....((((((((.....))))))))......)))))..)))))).))............--- ( -31.30, z-score = -2.16, R) >droAna3.scaffold_13266 10620964 99 + 19884421 --------GUUGGAGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAAGGCGA --------......(((.....((((----(((..((.(((((((((((....((((((((.....)))))))).....))).)).)))))))).)))))))....))).. ( -32.60, z-score = -1.34, R) >droMoj3.scaffold_6496 21709166 108 - 26866924 AAGUCGUGGCUUUUUUUUCUUCUCGUUUUCUUGCAAGCGCGUUAUCGCUGCGCAGCCGCAUAAAUAAUGCAGCUUAAAGAGCAGGUUGACGCCUACAAAAACAACGCC--- .......(((((.(((((.....((((..(((((..(((((.......)))))(((.((((.....)))).)))......)))))..)))).....))))).)).)))--- ( -29.10, z-score = -0.34, R) >droGri2.scaffold_15112 2727956 103 - 5172618 -----CUUGUGGAAGUCGUGGCGUUUUUGCUUGCAAGCGCGUUAUCGCUGCACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAACGAC--- -----.........(((((((((((...((((((..(((((....))).))..((((((((.....))))))))......)))))).))))))..........)))))--- ( -40.10, z-score = -3.08, R) >droWil1.scaffold_180701 2543491 98 - 3904529 ----------UCCUUUUUUUUCGCUUUGCUUUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGUGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAAGA--- ----------..(((((.(((.((((.((...))))))(((((((((((....((((((((.....)))))))).....))).)).))))))......))).))))).--- ( -27.10, z-score = -1.32, R) >droPer1.super_2 627131 98 + 9036312 --------GUUAAAGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUCCAAAAACAAGGCCUG- --------...........(((((((----(((..((.(((((((((((....((((((((.....)))))))).....))).)).)))))))).)))))))...)))..- ( -33.10, z-score = -1.90, R) >dp4.chr3 13215005 98 - 19779522 --------GUUCAAGUCGUGGCGUUU----UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUCCAAAAACAAGGCCUG- --------...........(((((((----(((..((.(((((((((((....((((((((.....)))))))).....))).)).)))))))).)))))))...)))..- ( -33.10, z-score = -1.83, R) >consensus _____________AGUCGUGGCGUUU____UUGCAAGCGCGUUAUCGCUGAACAGCCGCAUAAAUAAUGCGGCUUAAAGAGCAGGUUGACGCCUACAAAAACAAAGGG___ ...................((((((.....((((.((((......))))....((((((((.....))))))))......))))...)))))).................. (-25.98 = -26.72 + 0.74)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:31 2011