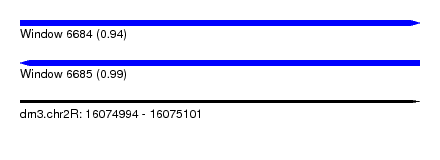
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,074,994 – 16,075,101 |
| Length | 107 |
| Max. P | 0.987796 |

| Location | 16,074,994 – 16,075,101 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 73.45 |
| Shannon entropy | 0.46666 |
| G+C content | 0.58233 |
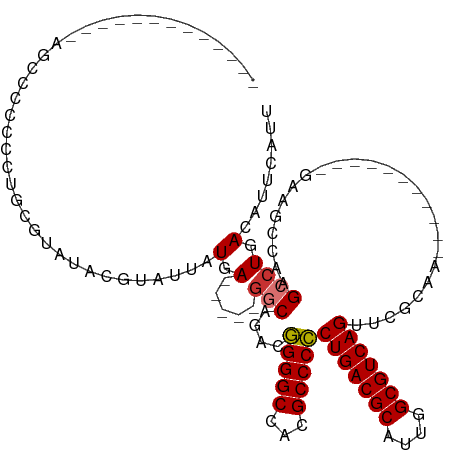
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -26.36 |
| Energy contribution | -25.82 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
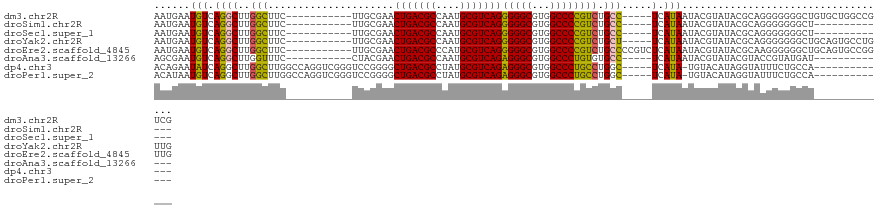
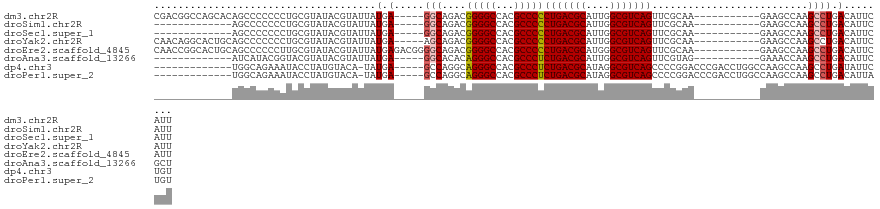
>dm3.chr2R 16074994 107 + 21146708 AAUGAAUGUCAGGCUUGGCUUC-----------UUGCGAACUGACGCCAAUGCGUCAGGGGGCGUGGCCCCGUCUGCC-----UCAUAAUACGUAUACGCAGGGGGGGCUGUGCUGGCCGUCG ...........(((..((((..-----------.......(((((((....)))))))..((((..(((((.(((((.-----..(((.....)))..))))).)))))..))))))))))). ( -46.80, z-score = -1.84, R) >droSim1.chr2R 14732824 94 + 19596830 AAUGAAUGUCAGGCUUGGCUUC-----------UUGCGAACUGACGCCAAUGCGUCAGGGGGCGUGGCCCCGUCUGCC-----UCAUAAUACGUAUACGCAGGGGGGGCU------------- .........((.((((.((...-----------..))...(((((((....))))))).)))).))(((((.(((((.-----..(((.....)))..))))).))))).------------- ( -38.30, z-score = -1.52, R) >droSec1.super_1 13635843 94 + 14215200 AAUGAAUGUCAGGCUUGGCUUC-----------UUGCGAACUGACGCCAAUGCGUCAGGGGGCGUGGCCCCGUCUGCC-----UCAUAAUACGUAUACGCAGGGGGGGCU------------- .........((.((((.((...-----------..))...(((((((....))))))).)))).))(((((.(((((.-----..(((.....)))..))))).))))).------------- ( -38.30, z-score = -1.52, R) >droYak2.chr2R 8008920 107 - 21139217 AAUGAAUGUCAGGCUUGGCUUC-----------UUGCGAACUGACGCCAAUGCGUCAGGGGGCGUGGCCCCGUCUGCU-----UCAUAAUACGUAUACGCAGGGGGGGCUGCAGUGCCUGUUG .........(((((...(((((-----------.......(((((((....))))))))))))(..(((((.(((((.-----...............))))).)))))..)...)))))... ( -46.80, z-score = -2.44, R) >droEre2.scaffold_4845 10225567 112 + 22589142 AAUGAAUGUCAGGCUUGGCUUC-----------UUGCGAACUGACGCCCAUGCGUCAGGGGGCGUGGCCCCGUCUGCCCCGUCUCAUAAUACGUAUACGCAAGGGGGGCUGCAGUGCCGGUUG ...........((((...((((-----------(((((..(((((((....)))))))((((((.(((...))))))))).................)))))))))(((......))))))). ( -44.00, z-score = -0.34, R) >droAna3.scaffold_13266 10618726 94 - 19884421 AGCGAAUGUCAGGCUUGGUUUC-----------CUACGAACUGACGCCAAUGCGUCAGAGGGCGUGGCCCUGUGUGCC-----UCAUAAUACGUAUACGUACCGUAUGAU------------- .(((..(((.((((........-----------.......(((((((....)))))))(((((...)))))....)))-----).))).((((....)))).))).....------------- ( -29.80, z-score = -0.78, R) >dp4.chr3 13212817 104 + 19779522 ACAGAAUAUCAGGCUUGGCUUGGCCAGGUCGGGUCCGGGGCUGACGCCUAUGCGUCAGAGGGCGUGGCCCUGCCUGGC-----UCAUA-UGUACAUAGGUAUUUCUGCCA------------- .(((((((((.((((((((...))))))))(((.(((((.(((((((....)))))))(((((...))))).))))))-----))...-........)))).)))))...------------- ( -49.20, z-score = -2.63, R) >droPer1.super_2 624981 104 - 9036312 ACAUAAUGUCAGGCUUGGCUUGGCCAGGUCGGGUCCGGGGCUGACGCCUAUGCGUCAGAGGGCGUGGCCCUGCCUGGC-----UCAUA-UGUACAUAGGUAUUUCUGCCA------------- (((((..(((((((.((((...))))(((((.((((....(((((((....))))))).)))).)))))..)))))))-----...))-))).....((((....)))).------------- ( -49.80, z-score = -2.74, R) >consensus AAUGAAUGUCAGGCUUGGCUUC___________UUGCGAACUGACGCCAAUGCGUCAGGGGGCGUGGCCCCGUCUGCC_____UCAUAAUACGUAUACGCAGGGGGGGCU_____________ .........(((((..........................(((((((....)))))))(((((...))))))))))............................................... (-26.36 = -25.82 + -0.53)
| Location | 16,074,994 – 16,075,101 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 73.45 |
| Shannon entropy | 0.46666 |
| G+C content | 0.58233 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -21.97 |
| Energy contribution | -21.87 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
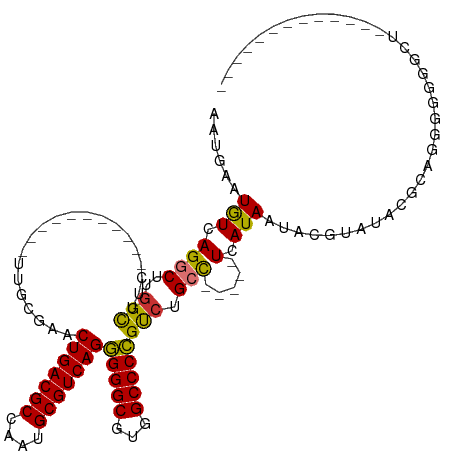
>dm3.chr2R 16074994 107 - 21146708 CGACGGCCAGCACAGCCCCCCCUGCGUAUACGUAUUAUGA-----GGCAGACGGGGCCACGCCCCCUGACGCAUUGGCGUCAGUUCGCAA-----------GAAGCCAAGCCUGACAUUCAUU ....(((..((...(((((..((((.((((.....)))).-----.))))..)))))........(((((((....)))))))(((....-----------)))))...)))........... ( -37.30, z-score = -1.84, R) >droSim1.chr2R 14732824 94 - 19596830 -------------AGCCCCCCCUGCGUAUACGUAUUAUGA-----GGCAGACGGGGCCACGCCCCCUGACGCAUUGGCGUCAGUUCGCAA-----------GAAGCCAAGCCUGACAUUCAUU -------------.(((((..((((.((((.....)))).-----.))))..)))))((.((...(((((((....)))))))(((....-----------))).....)).))......... ( -33.30, z-score = -2.35, R) >droSec1.super_1 13635843 94 - 14215200 -------------AGCCCCCCCUGCGUAUACGUAUUAUGA-----GGCAGACGGGGCCACGCCCCCUGACGCAUUGGCGUCAGUUCGCAA-----------GAAGCCAAGCCUGACAUUCAUU -------------.(((((..((((.((((.....)))).-----.))))..)))))((.((...(((((((....)))))))(((....-----------))).....)).))......... ( -33.30, z-score = -2.35, R) >droYak2.chr2R 8008920 107 + 21139217 CAACAGGCACUGCAGCCCCCCCUGCGUAUACGUAUUAUGA-----AGCAGACGGGGCCACGCCCCCUGACGCAUUGGCGUCAGUUCGCAA-----------GAAGCCAAGCCUGACAUUCAUU ...(((((...((.(((((..((((.((((.....)))).-----.))))..)))))........(((((((....)))))))(((....-----------)))))...)))))......... ( -41.00, z-score = -3.66, R) >droEre2.scaffold_4845 10225567 112 - 22589142 CAACCGGCACUGCAGCCCCCCUUGCGUAUACGUAUUAUGAGACGGGGCAGACGGGGCCACGCCCCCUGACGCAUGGGCGUCAGUUCGCAA-----------GAAGCCAAGCCUGACAUUCAUU .....(((...((.(((((.....((((.......))))....)))))....(((((...)))))(((((((....)))))))(((....-----------)))))...)))........... ( -42.20, z-score = -1.65, R) >droAna3.scaffold_13266 10618726 94 + 19884421 -------------AUCAUACGGUACGUAUACGUAUUAUGA-----GGCACACAGGGCCACGCCCUCUGACGCAUUGGCGUCAGUUCGUAG-----------GAAACCAAGCCUGACAUUCGCU -------------.(((((..(((((....))))))))))-----(((..(((((((...)))))(((((((....)))))))...)).(-----------....)...)))........... ( -31.00, z-score = -2.10, R) >dp4.chr3 13212817 104 - 19779522 -------------UGGCAGAAAUACCUAUGUACA-UAUGA-----GCCAGGCAGGGCCACGCCCUCUGACGCAUAGGCGUCAGCCCCGGACCCGACCUGGCCAAGCCAAGCCUGAUAUUCUGU -------------((((.....(((....)))..-..((.-----((((((..(((((.......(((((((....)))))))....)).)))..)))))))).))))............... ( -38.00, z-score = -1.92, R) >droPer1.super_2 624981 104 + 9036312 -------------UGGCAGAAAUACCUAUGUACA-UAUGA-----GCCAGGCAGGGCCACGCCCUCUGACGCAUAGGCGUCAGCCCCGGACCCGACCUGGCCAAGCCAAGCCUGACAUUAUGU -------------((((.....(((....)))..-..((.-----((((((..(((((.......(((((((....)))))))....)).)))..)))))))).))))............... ( -38.00, z-score = -2.09, R) >consensus _____________AGCCCCCCCUGCGUAUACGUAUUAUGA_____GGCAGACGGGGCCACGCCCCCUGACGCAUUGGCGUCAGUUCGCAA___________GAAGCCAAGCCUGACAUUCAUU .............................................(((....(((((...)))))(((((((....)))))))..........................)))........... (-21.97 = -21.87 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:28 2011