| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,069,399 – 16,069,490 |
| Length | 91 |
| Max. P | 0.994902 |

| Location | 16,069,399 – 16,069,490 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.60768 |
| G+C content | 0.42255 |
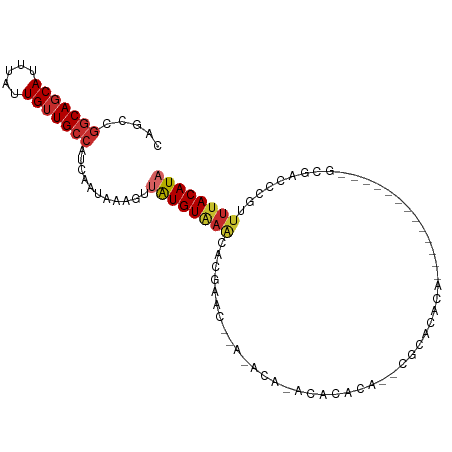
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -10.16 |
| Energy contribution | -10.58 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
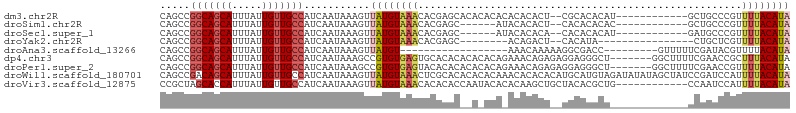
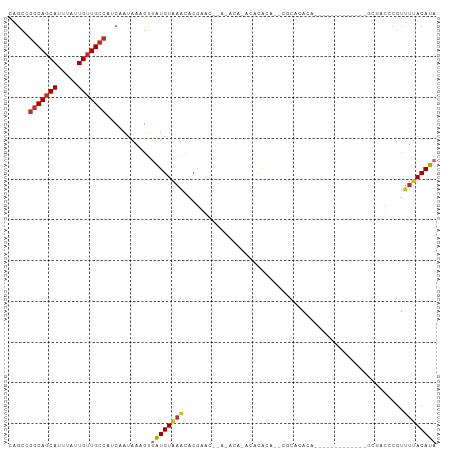
>dm3.chr2R 16069399 91 - 21146708 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACGAGCACACACACACACACU--CGCACACAU------------GCUGCCCGUUUUACAUA .....(((((((.....)))))))...........((((((((.(((.(((..............--.(((....)------------))))).))))))))))) ( -22.06, z-score = -2.46, R) >droSim1.chr2R 14725004 85 - 19596830 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACGAGC------AUACACACU--CACACACAC------------GCUGCCCGUUUUACAUA .....(((((((.....)))))))...........((((((((.(((.((------(........--.........------------..))).))))))))))) ( -21.01, z-score = -3.00, R) >droSec1.super_1 13630398 85 - 14215200 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACGAGC------AUACACACA--CACACACAU------------GAUGCCCGUUUUACAUA .....(((((((.....)))))))...........((((((((.(((.((------((.((....--........)------------))))).))))))))))) ( -23.10, z-score = -3.90, R) >droYak2.chr2R 8003167 79 + 21139217 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACGAGC--------ACAGACU--CACAUA----------------CUGCUCGUUUUACAUA .....(((((((.....)))))))...........((((((((.((((((--------(......--......----------------.))))))))))))))) ( -26.12, z-score = -5.34, R) >droAna3.scaffold_13266 10613708 78 + 19884421 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGU------------------AAACAAAAAGGCGACC---------GUUUUUCGAUACGUUUUACAUA ..((((((((((.....))))))).........(((....------------------.))).....)))...(---------((........)))......... ( -18.00, z-score = -2.15, R) >dp4.chr3 13207399 98 - 19779522 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGCCGUGUGAGUGCACACACACACAGAAACAGAGAGGAGGGCU-------GGCUUUUCGAACCGCUUUACAUA .....(((((((.....))))))).....((((((.(((((.((....)).))))).........((((((....-------..)))))).....)))))).... ( -29.70, z-score = -2.02, R) >droPer1.super_2 619577 98 + 9036312 CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGCCGUGUGAGUACACACACACACAGAAACAGAGAGGAGGGCU-------GGCUUUUCGAACCGUUUUACAUA .(((((((((((.....)))))))........(((((((((.((....)).))))).....(.....)...))))-------))))................... ( -28.50, z-score = -2.60, R) >droWil1.scaffold_180701 2525867 105 - 3904529 CAGCCGACAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACUCGCACACACACAAACACACACAUGCAUGUAGAUAUAUAGCUAUCCGAUCCAUUUUACAUA .....(.(((((.....))))).)........(((((((((.((..(((..................)))..))...)))))))))................... ( -11.97, z-score = 0.62, R) >droVir3.scaffold_12875 17185862 93 + 20611582 CCGCUAGCACCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACACACCAAUACACACAAGCUGCUACACGCUG------------CCAAUCCAUUUUACAUA ..(((((((.(.(((((((.......)))))))(((.....)))..................).)))))...))..------------................. ( -9.60, z-score = 0.54, R) >consensus CAGCCGGCAGCAUUUAUUGUUGCCAUCAAUAAAGUUAUGUAAACACGAAC__A_ACA_ACACACA__CGCACACA_____________GCGACCCGUUUUACAUA .....(((((((.....)))))))...........((((((((......................................................)))))))) (-10.16 = -10.58 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:26 2011