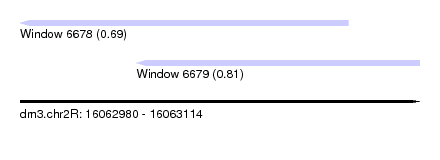
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,062,980 – 16,063,114 |
| Length | 134 |
| Max. P | 0.813870 |

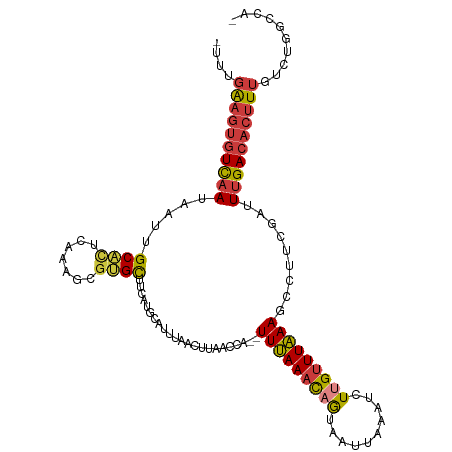
| Location | 16,062,980 – 16,063,090 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.37934 |
| G+C content | 0.33667 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.06 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16062980 110 - 21146708 -UUUGAAGUGUCAAUAAUUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCA--UUUAAACAGUAAUUAAAUCUUGUUUAAAACCUUCGAUUUGACACUUUGCCUGGCCA- -...((((((((((..((((........(((((...)))))..............--(((((((((..........))))))))).....)))))))))))))).........- ( -21.20, z-score = -1.65, R) >droSim1.chr2R 14718817 108 - 19596830 ---UGAAGUGUCAAUAAGUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCA--UUUAAACAGUAAUUAAAUCUUGUUUAAAGCCUUCGAUUUGACACUUUGUCUGGCCA- ---.((((((((((.(((.((.......(((((...)))))..............--(((((((((..........))))))))))))))....)))))))))).........- ( -24.50, z-score = -2.03, R) >droSec1.super_1 13624247 108 - 14215200 ---UGAAGUGUCAAUAAGUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCA--UUUAAACAGUAAUUAAAUCUUGUUUAAAGCCUUCGAUUUGACACUUUGUCUGGCCA- ---.((((((((((.(((.((.......(((((...)))))..............--(((((((((..........))))))))))))))....)))))))))).........- ( -24.50, z-score = -2.03, R) >droYak2.chr2R 7996878 110 + 21139217 -UUUGAAGUGUCAAUAAUUGCACUCGAAGCGUGCUUCAUGCAUUUAACUUAACCA--UUUAAACAGUAAUUAAAUCUUGUUUAAAGCCUUCGAUUUGACACUUUGUCUGGCCA- -...((((((((((.........(((((((((((.....))))............--(((((((((..........)))))))))).)))))).)))))))))).........- ( -24.90, z-score = -2.30, R) >droEre2.scaffold_4845 10212513 110 - 22589142 -UUUGAAGUGUCAAUAAUUGCACUCAAAGCGCGCUUCAUGCAUUCAACUUAACCA--UUUAAACAGUAAUUAAAUCUUGUUUAAAGCCUUCGAUUUGACACUUUGUCUGGCCA- -...((((((((((..((((......(((.((((.....))..............--(((((((((..........)))))))))))))))))))))))))))).........- ( -20.90, z-score = -1.06, R) >droAna3.scaffold_13266 10606544 103 + 19884421 UUUUGGUGUGUCCAUAAUUGCA---------UGUUUCGUGCAUUUGAGUUGGCCA--UUUAAAUUGUAAUUAAAUUUUGCUUAAAGCCUUCAAAUUGACACUUUGACUGGCCAA ...(((.....)))(((.((((---------((...)))))).)))..(((((((--.(((((.(((.....(((((.((.....))....))))).))).))))).))))))) ( -23.50, z-score = -1.21, R) >droWil1.scaffold_180701 2508456 112 - 3904529 -AUUCAACAAUUAAUAAUUGCUUUAACCUUGAGUCGCAUUUAAUGCAUUUUGUGAAUUUCAAAUUAUAAUUAAAUUUCGUUUGAAGCAUUAAAAUUGACACUUUGACUGGCCG- -..................(((........(.((((..((((((((...((((((........))))))((((((...)))))).))))))))..)))).).......)))..- ( -14.46, z-score = 0.50, R) >consensus _UUUGAAGUGUCAAUAAUUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCA__UUUAAACAGUAAUUAAAUCUUGUUUAAAGCCUUCGAUUUGACACUUUGUCUGGCCA_ ....((((((((((.....((((.......)))).......................(((((((((..........))))))))).........)))))))))).......... (-14.91 = -15.06 + 0.15)

| Location | 16,063,019 – 16,063,114 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.81 |
| Shannon entropy | 0.50050 |
| G+C content | 0.33786 |
| Mean single sequence MFE | -15.83 |
| Consensus MFE | -7.83 |
| Energy contribution | -7.92 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16063019 95 - 21146708 AAUCCUCCAAGCUUACCAUUACUCUUUGAAGUGUCAAUAAUUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCAUUUAAACAGUAAUUAAA .................((((((((((((.((((........)))))))))).((((.....))))...................)))))).... ( -14.40, z-score = -1.92, R) >droSim1.chr2R 14718856 83 - 19596830 ------------AAUCCUCCAAGCUUUGAAGUGUCAAUAAGUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCAUUUAAACAGUAAUUAAA ------------..........(((((((.((((........)))))))))))((((.....))))............................. ( -16.40, z-score = -2.55, R) >droSec1.super_1 13624286 83 - 14215200 ------------AAUCCGCCAAGCUUUGAAGUGUCAAUAAGUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCAUUUAAACAGUAAUUAAA ------------..........(((((((.((((........)))))))))))((((.....))))............................. ( -16.40, z-score = -1.98, R) >droYak2.chr2R 7996917 95 + 21139217 AAUACUACCAGUUUACCACUACUCUUUGAAGUGUCAAUAAUUGCACUCGAAGCGUGCUUCAUGCAUUUAACUUAACCAUUUAAACAGUAAUUAAA ..((((....(((((.....((.((((((.((((........)))))))))).))((.....))................)))))))))...... ( -14.70, z-score = -1.40, R) >droEre2.scaffold_4845 10212552 95 - 22589142 AAUCCACCAAGCUUACCAUUACACUUUGAAGUGUCAAUAAUUGCACUCAAAGCGCGCUUCAUGCAUUCAACUUAACCAUUUAAACAGUAAUUAAA .................(((((.((((((.((((........))))))))))...((.....))......................))))).... ( -13.30, z-score = -1.49, R) >droAna3.scaffold_13266 10606584 94 + 19884421 UGACCAUUAAAUUUUCCGGCACAUGAGAUUCUUUUGGUGUGUCCA-UAAUUGCAUGUUUCGUGCAUUUGAGUUGGCCAUUUAAAUUGUAAUUAAA ......((((((...(((((((((.(((....))).)))))))..-(((.((((((...)))))).)))....))..))))))............ ( -19.80, z-score = -0.74, R) >consensus AAUCC__CAAG_UUACCACCACACUUUGAAGUGUCAAUAAGUGCACUCAAAGCGUGCUUCAUGCAUUUAACUUAACCAUUUAAACAGUAAUUAAA .......................((((((.((((........)))))))))).((((.....))))............................. ( -7.83 = -7.92 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:24 2011