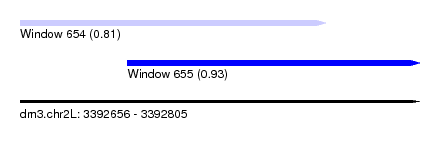
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,392,656 – 3,392,805 |
| Length | 149 |
| Max. P | 0.931764 |

| Location | 3,392,656 – 3,392,770 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Shannon entropy | 0.36215 |
| G+C content | 0.52829 |
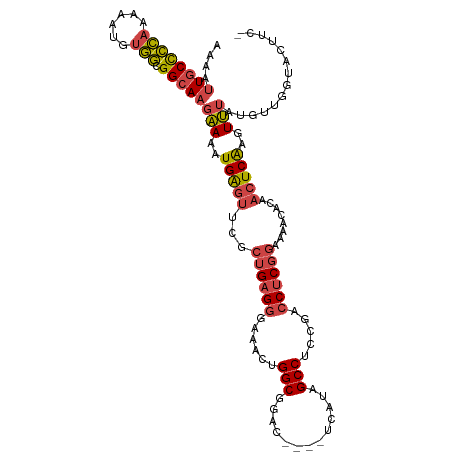
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -18.37 |
| Energy contribution | -19.99 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
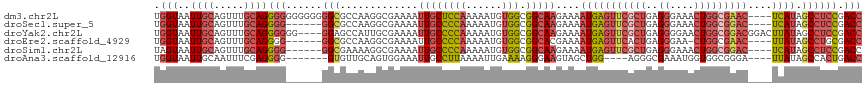
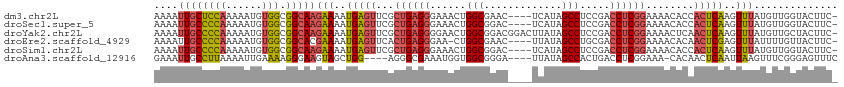
>dm3.chr2L 3392656 114 + 23011544 UGGUAAUUGCAGUUUGCAGGGGGGGGGGGGCGCCAAGGCGAAAAUUGCUCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGAAC----UCAUAGCCUCCGACC ......((((.....))))..((..((((((((....)).....((((((((......))).)))))....(((((((((((..((....)))))))))----)))).))))))..)) ( -47.10, z-score = -3.58, R) >droSec1.super_5 1539445 108 + 5866729 UGGUAAUUGCAGUUUGCAGGGG------GGCGCCAAGGCGAAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC----UCAUAGCCUCCGACC .(((..((((.....))))(((------(((((....)).....((((((((......))).)))))....(((((((((((..((....)))))))))----)))).)))))).))) ( -48.60, z-score = -4.23, R) >droYak2.chr2L 3377715 114 + 22324452 UGGUAAUUGCAGUUUGCAGGGGGG----GGAGCCAUUGCGAAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGGAACUGGCGGACGGACUUAUAGCCUCCGACC .(((...(((.....)))((((((----....))...((.....((((((((......))).)))))....((((((((((((.((....))..))).))))))))).)))))).))) ( -39.10, z-score = -1.16, R) >droEre2.scaffold_4929 3422325 107 + 26641161 UGGUAAUUGCAGUUUGCAGGGG------GGCGCCAAGGCGAAAAUUGCCCCAAAAAUGUGGCGGCACGAAAAUGAGUUCACUGAGGGAA-CUGGCGAAC----UUAUAGCCUGCGACC .(((..((((.....))))(.(------((((((..(((((...)))))(((......))).)))......((((((((.((.((....-)))).))))----)))).)))).).))) ( -36.10, z-score = -1.15, R) >droSim1.chr2L 3331141 108 + 22036055 UAGUAAUUGCAGUUUGCAGGGG------GGCGAAAAGGCGAAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC----UCAUAGCCUCCGACC ......((((.....))))(((------(((.............((((((((......))).)))))....(((((((((((..((....)))))))))----)))).)))))).... ( -44.80, z-score = -4.30, R) >droAna3.scaffold_12916 2854333 103 - 16180835 UGGUAAUUGCAAUUUCGAGGGG-------GUGUUGCAGUGGAAAUUGCCUUAAAAUUGAAAAGGGAAGUAGCUGG----AGGGCGAAAUGGUGGCGGGA----UUAUAGCCACUGACC .(((.((((((((.((.....)-------).)))))))).....(((((((........................----)))))))...((((((....----.....)))))).))) ( -24.46, z-score = -0.64, R) >consensus UGGUAAUUGCAGUUUGCAGGGG______GGCGCCAAGGCGAAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC____UCAUAGCCUCCGACC .(((...(((.....)))((((........(((....))).......))))........((.(((..........(((((((..((....))))))))).........))).)).))) (-18.37 = -19.99 + 1.62)
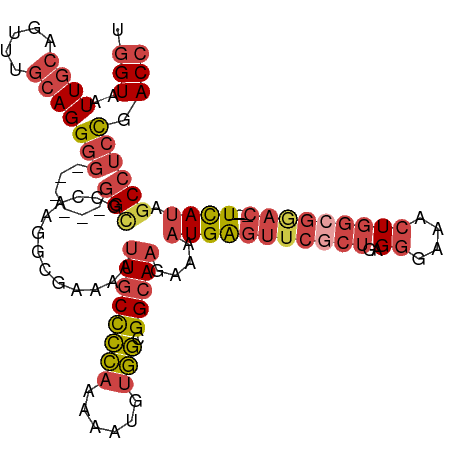
| Location | 3,392,696 – 3,392,805 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Shannon entropy | 0.33736 |
| G+C content | 0.46380 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.64 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3392696 109 + 23011544 AAAAUUGCUCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGAAC----UCAUAGCCUCCGACCUCGGAAAACACCACUCAAGUUUAUGUUGGUACUUC- ....((((((((......))).)))))....(((((((((((..((....)))))))))----)))).(((((((....)))).((((..((....))...))))))).....- ( -35.00, z-score = -3.13, R) >droSec1.super_5 1539479 109 + 5866729 AAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC----UCAUAGCCUCCGACCUCGGAAAACACCACUCAAGUUUAUGUUGGUACUUC- ....((((((((......))).)))))....(((((((((((..((....)))))))))----)))).(((((((....)))).((((..((....))...))))))).....- ( -37.50, z-score = -3.32, R) >droYak2.chr2L 3377751 113 + 22324452 AAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGGAACUGGCGGACGGACUUAUAGCCUCCGACCUCGGAAAACUCAACUCAAGUUUAUGUUGCUACUUC- .................(((((((((.(((..((((((..((((((........((((.((........)))))).))))))..)))))).......))).)))))))))...- ( -34.60, z-score = -2.02, R) >droEre2.scaffold_4929 3422359 108 + 26641161 AAAAUUGCCCCAAAAAUGUGGCGGCACGAAAAUGAGUUCACUGAGGGAA-CUGGCGAAC----UUAUAGCCUGCGACCUCGGAAAACACAACUCGAGUUUAUUUUGUUACUUC- .....(((((((......))).))))(((...((.(((..((((((...-..(((....----.....))).....))))))..))).))..)))..................- ( -27.70, z-score = -1.52, R) >droSim1.chr2L 3331175 109 + 22036055 AAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC----UCAUAGCCUCCGACCUCGGAAAACACCACUCAAGUUUAUGUUGGUACUUC- ....((((((((......))).)))))....(((((((((((..((....)))))))))----)))).(((((((....)))).((((..((....))...))))))).....- ( -37.50, z-score = -3.32, R) >droAna3.scaffold_12916 2854366 105 - 16180835 GAAAUUGCCUUAAAAUUGAAAAGGGAAGUAGCUGG----AGGGCGAAAUGGUGGCGGGA----UUAUAGCCACUGACCUCGGAAA-CACAACUCAAUUAAGUUUCGGGAGUUUC ((((((.(((.(((.((((...(((..((.....(----(((.......((((((....----.....))))))..))))(....-)))..)))..)))).))).))))))))) ( -25.70, z-score = -0.78, R) >consensus AAAAUUGCCCCAAAAAUGUGGCGGCAAGAAAAUGAGUUCGCUGAGGGAAACUGGCGGAC____UCAUAGCCUCCGACCUCGGAAAACACAACUCAAGUUUAUGUUGGUACUUC_ ....((((((((......))).)))))(((..(((((...((((((......(((.............))).....))))))........)))))..))).............. (-20.22 = -20.64 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:45 2011