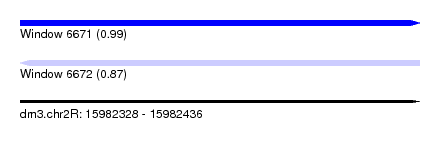
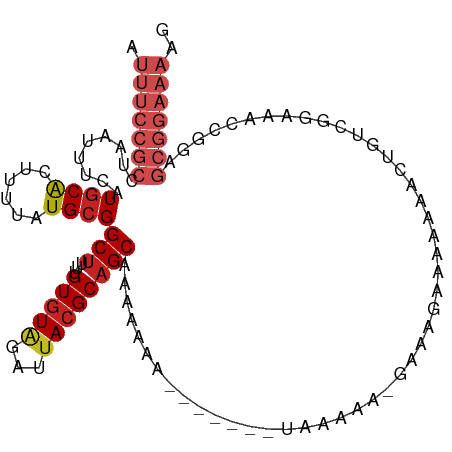
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,982,328 – 15,982,436 |
| Length | 108 |
| Max. P | 0.988766 |

| Location | 15,982,328 – 15,982,436 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.59370 |
| G+C content | 0.38855 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -13.08 |
| Energy contribution | -14.95 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15982328 108 + 21146708 CUUUUCCGCUCCGGUUUCCGACAGUUUUUUUUUUUUUUUCUUA--CCACAUUUUUUUGCUGCGUAAUCUACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..((((((....)...................--...(((((((.(((.((...............)).))).)))))))...)))))..)))))))). ( -23.26, z-score = -1.87, R) >droSim1.chr2R 14629975 102 + 19596830 CUUUUCCGCUCCGGUUUCCGACAGUUUUUUUUU------CUUA--CCACAUUUUUUUGCUGCGUAAUCUACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.((.((.....)).)------)...--...(((((((.(((.((...............)).))).)))))))...)))))..)))))))). ( -24.06, z-score = -2.16, R) >droSec1.super_1 13457377 102 + 14215200 CUUUUCCGCUCCGGUUUCCGACAGUUUUUUUUU------CUUA--CCACAUUUUUUUGCUGCGUAAUCUACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.((.((.....)).)------)...--...(((((((.(((.((...............)).))).)))))))...)))))..)))))))). ( -24.06, z-score = -2.16, R) >droYak2.chr2R 7913255 110 - 21139217 CUUUUCCGCUCCGGUUUCCGACAGCUUUUUUCUUACCACAUUUAUUUUUAUUUUUUUGCUGCGUAAUCAACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((((.((((................................)))))).................((((....))))..)))))..)))))))). ( -24.35, z-score = -2.05, R) >droEre2.scaffold_4845 10130265 99 + 22589142 CUUUUCCGCUCCGGUUUCCGACAGCUUUUUUCUUACCACAUUU-----------UUUGCUGCGUAAUCCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.....((((((...((((((((...-----------..)).)).)))).......)))))).((((....))))..)))))..)))))))). ( -24.90, z-score = -2.25, R) >droAna3.scaffold_13266 10526376 110 - 19884421 UUUUUCCGCUCCGGUUUCUUUCGGUUUUUUCUUACCAUUCUCAUAAAUUUUUUUUGUGCAGCGUAAUCUACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((....(((((((..........((((((((.....)))))).)).(((...)))..)))))))((((....))))..)))))..)))))))). ( -28.70, z-score = -3.52, R) >dp4.chr3 13117056 100 + 19779522 UUUUUCCGCUCCGGUUUCC----GUAUAUUGCUUUCGUUUCAACA------UUUUCUGCUGCGUAAUCCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.----.......((((((((..((...------.....))..)))...........))))).((((....))))..)))))..)))))))). ( -23.00, z-score = -1.41, R) >droPer1.super_2 529423 100 - 9036312 UUUUUCCGCUCCGGUUUCC----GUAUAUUGCUUUCGUUUCAACA------UUUUCUGCUGCGUAAUCCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.----.......((((((((..((...------.....))..)))...........))))).((((....))))..)))))..)))))))). ( -23.00, z-score = -1.41, R) >droWil1.scaffold_180701 2363020 93 + 3904529 -------------GUUUUC----UGCUUUGCGUUUCCUCACACGGAGCAUUUUUUUCGCUGCGUAAUUUAGACAAAAGCCGCAUAAAAGCGUAUGAAAUUAGACGGAAAU -------------..((((----((((((..((((......(((.(((.........))).))).....)))).)))))(((......))).............))))). ( -17.80, z-score = 0.53, R) >droVir3.scaffold_12875 17058069 86 - 20611582 ------------------------UUUUUCCGCUUCGCUUUAAAAGCCAUUUUUUUCGCUGCGUAAUUCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ------------------------..(((((((((.(((.....)))(((((((...((.((...............)).))..))))))).........))))))))). ( -19.66, z-score = -1.13, R) >droMoj3.scaffold_6496 21558432 98 + 26866924 UUUUUCCGCUUUGGUUUCAGAGUUGUUAUUUCU------UCAAU------UUUUUUCGCUGCGUAAUUCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((((.(((((((((((((........------.))))------)).....(((..((.......))...))).((((....)))).)))))))))))))))). ( -23.70, z-score = -1.93, R) >consensus CUUUUCCGCUCCGGUUUCCGACAGUUUUUUUCUUUC_UUCUAA_______UUUUUUUGCUGCGUAAUCCACACAAAAGCCGCAUAAAAGUGCAUGAAAUUAGGCGGAAAU ..(((((((.(..(((((.......................................(((..((.......))...))).((((....))))..)))))..)))))))). (-13.08 = -14.95 + 1.87)

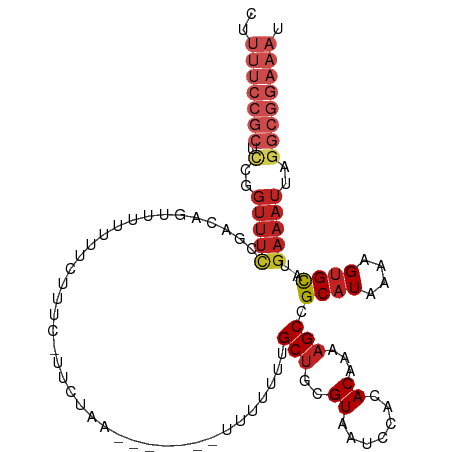
| Location | 15,982,328 – 15,982,436 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.59370 |
| G+C content | 0.38855 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -11.69 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15982328 108 - 21146708 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUAGAUUACGCAGCAAAAAAAUGUGG--UAAGAAAAAAAAAAAAAAACUGUCGGAAACCGGAGCGGAAAAG .(((((((((..((((..(((......)))(((...((((((...))))))(((......)))..--.....................))).))))..)).))))))).. ( -22.60, z-score = -0.66, R) >droSim1.chr2R 14629975 102 - 19596830 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUAGAUUACGCAGCAAAAAAAUGUGG--UAAG------AAAAAAAAACUGUCGGAAACCGGAGCGGAAAAG .(((((((((..((((..(((......)))(((...((((((...))))))(((......)))..--....------...........))).))))..)).))))))).. ( -22.60, z-score = -0.56, R) >droSec1.super_1 13457377 102 - 14215200 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUAGAUUACGCAGCAAAAAAAUGUGG--UAAG------AAAAAAAAACUGUCGGAAACCGGAGCGGAAAAG .(((((((((..((((..(((......)))(((...((((((...))))))(((......)))..--....------...........))).))))..)).))))))).. ( -22.60, z-score = -0.56, R) >droYak2.chr2R 7913255 110 + 21139217 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUUGAUUACGCAGCAAAAAAAUAAAAAUAAAUGUGGUAAGAAAAAAGCUGUCGGAAACCGGAGCGGAAAAG .(((((((((..((((..(((......)))(((((((.(.(((....((((...................)))).))).).)))))))....))))..)).))))))).. ( -25.71, z-score = -1.18, R) >droEre2.scaffold_4845 10130265 99 - 22589142 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUGGAUUACGCAGCAAA-----------AAAUGUGGUAAGAAAAAAGCUGUCGGAAACCGGAGCGGAAAAG .(((((((((..((((..((((((((.(((.(((((((((((.....)))).))))-----------)...)).)))....))))).)))..))))..)).))))))).. ( -27.90, z-score = -1.56, R) >droAna3.scaffold_13266 10526376 110 + 19884421 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUAGAUUACGCUGCACAAAAAAAAUUUAUGAGAAUGGUAAGAAAAAACCGAAAGAAACCGGAGCGGAAAAA .((((((((((.(((((((((......)))...((((((((((......)))))))))).......)))))).)))..........(((........))).))))))).. ( -32.00, z-score = -3.92, R) >dp4.chr3 13117056 100 - 19779522 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUGGAUUACGCAGCAGAAAA------UGUUGAAACGAAAGCAAUAUAC----GGAAACCGGAGCGGAAAAA .(((((((((..((((.((((......))))(((((((((((.....)))((((.....------))))...)))))))).......----.))))..)).))))))).. ( -29.90, z-score = -2.67, R) >droPer1.super_2 529423 100 + 9036312 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUGGAUUACGCAGCAGAAAA------UGUUGAAACGAAAGCAAUAUAC----GGAAACCGGAGCGGAAAAA .(((((((((..((((.((((......))))(((((((((((.....)))((((.....------))))...)))))))).......----.))))..)).))))))).. ( -29.90, z-score = -2.67, R) >droWil1.scaffold_180701 2363020 93 - 3904529 AUUUCCGUCUAAUUUCAUACGCUUUUAUGCGGCUUUUGUCUAAAUUACGCAGCGAAAAAAAUGCUCCGUGUGAGGAAACGCAAAGCA----GAAAAC------------- ............((((...(((......)))(((((.((.....((((((((((.......))))..))))))(....)))))))).----))))..------------- ( -19.60, z-score = -0.38, R) >droVir3.scaffold_12875 17058069 86 + 20611582 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUGAAUUACGCAGCGAAAAAAAUGGCUUUUAAAGCGAAGCGGAAAAA------------------------ .(((((((....((((.(((...((((((((.....))))))))....)))..))))......(((.....)))...)))))))..------------------------ ( -21.00, z-score = -0.68, R) >droMoj3.scaffold_6496 21558432 98 - 26866924 AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUGAAUUACGCAGCGAAAAAA------AUUGA------AGAAAUAACAACUCUGAAACCAAAGCGGAAAAA .(((((((....(((((((((......))))..(((((((((.....)))).)))))...------.....------..............))))).....))))))).. ( -19.20, z-score = -0.90, R) >consensus AUUUCCGCCUAAUUUCAUGCACUUUUAUGCGGCUUUUGUGUAGAUUACGCAGCAAAAAAA_______UAAAAA_GAAAGAAAAAAACUGUCGGAAACCGGAGCGGAAAAG .(((((((.........((((......))))(((...(((((...))))))))................................................))))))).. (-11.69 = -13.04 + 1.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:18 2011