| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,979,611 – 15,979,705 |
| Length | 94 |
| Max. P | 0.927486 |

| Location | 15,979,611 – 15,979,705 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.31 |
| Shannon entropy | 0.52155 |
| G+C content | 0.50696 |
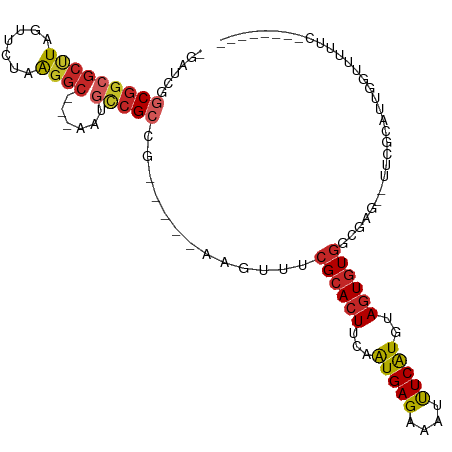
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
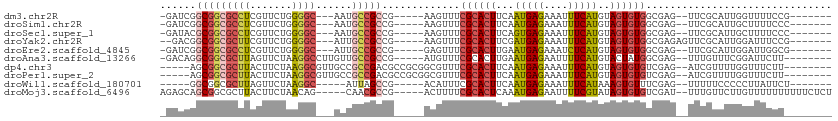
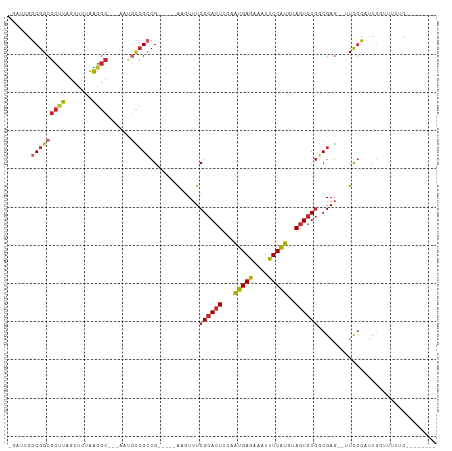
>dm3.chr2R 15979611 94 + 21146708 -GAUCGGCGGCGCCUCGUUCUGGGGC---AAUGCCGCCG-----AAGUUUCGCACUUCAAUGAGAAAUUUCAUGUAGUGUGGCGAG--UUCGCAUUGGUUUUCCG------- -..(((((((((((((.....)))))---...)))))))-----).....((((((...(((((....)))))..))))))(((..--..)))...((....)).------- ( -39.40, z-score = -3.23, R) >droSim1.chr2R 14627287 94 + 19596830 -GAUCGGCGGCGCCUCGUUCUGGGGC---AAUGCCGCCG-----AAGUUUCGCACUUCAAUGAGAAAUUUCAUGUAGUGUGGCGAG--UUCGCAUUGCUUUUCCC------- -..(((((((((((((.....)))))---...)))))))-----).....((((((...(((((....)))))..))))))(((..--..)))............------- ( -38.50, z-score = -2.99, R) >droSec1.super_1 13454673 94 + 14215200 -GAUACGCGGCGCCUCGUUCUGGGGC---AAUGCCGCCG-----AAGUUUCGCACUUCAGUGAGAAAUUUCAUGUAGUGUGGCGAG--UUCGCAUUGCUUUUCCC------- -.....((((((((((.....)))))---...))))).(-----(((((((.(((....))).))))))))..(((((((((....--.))))))))).......------- ( -37.00, z-score = -2.72, R) >droYak2.chr2R 7910435 95 - 21139217 --GACGGCGGCGCUUCGUUCUGGGGC---AUUGCCGCCG-----AAGUUUCGCACUUCGAUGAGAAAUUUCAUGUAGUGUGGCGAGAGUUCGCAUUGGAUUUCCG------- --...(((((((((((.....)))))---...))))))(-----(((((((.((......)).))))))))((.(((((((((....).)))))))).)).....------- ( -35.20, z-score = -1.38, R) >droEre2.scaffold_4845 10127598 94 + 22589142 -GAUCGGCGGCGCCUCGUUCUGGGGC---AUUGCCGCCG-----GAGUUUCGCACUUGAAUGAGAAAUCUCAUGUAGUGUGGCGAG--UUCGCAUUGGAUUGGCG------- -..(((((((((((((.....)))))---...)))))))-----).....((((((...(((((....)))))..))))))((.((--(((.....))))).)).------- ( -43.30, z-score = -3.38, R) >droAna3.scaffold_13266 10523518 96 - 19884421 -GACAGGCGGCGCUUAGUUCUAAGGCCUUGUUGCCGCCG-----AUGUUUCGCACUUGAAUGAGAAUUUUCAUGUACUAUGGCGAG--UUUGUUUCGGAUUCUU-------- -((((((((((.((((....)))))))...(((((((.(-----(....))))((.((((........)))).)).....))))))--))))))..........-------- ( -25.10, z-score = -0.06, R) >dp4.chr3 13113887 97 + 19779522 -----AGCGGCGCUUACUUCUAAGGCGUUGCCGCCGACGCCGCGGCGUUUCGCACUUCAAUGAGAAAUUUCAUGUAGUGUGUCGAG--AUCGUUUUGGUUUCUU-------- -----.(((((((((.......)))))))))(((((......)))))...((((((...(((((....)))))..))))))..(((--(((.....))))))..-------- ( -34.40, z-score = -1.61, R) >droPer1.super_2 526140 97 - 9036312 -----AGCGGCGCUUACUUCUAAGGCGUUGCCGCCGACGCCGCGGCGUUUCGCACUUCAAUGAGAAAUUUCAUGUAGUGUGUCGAG--AUCGUUUUGGUUUCUU-------- -----.(((((((((.......)))))))))(((((......)))))...((((((...(((((....)))))..))))))..(((--(((.....))))))..-------- ( -34.40, z-score = -1.61, R) >droWil1.scaffold_180701 2354772 88 + 3904529 -----GGCGGCGCUUAGUUCUAAGGC-----AUUAGCCG-----ACAUUUCGCACUUCAAUGAGAAAUUUCAUAAAGUGUUUCGAG--UUUUUCCCCCUUAUUCU------- -----(((.((.((((....))))))-----....)))(-----((.....((((((..(((((....))))).)))))).....)--))...............------- ( -17.40, z-score = -0.49, R) >droMoj3.scaffold_6496 21552950 100 + 26866924 AGAGCAGCGGCGCUUACUUCUAACAG-----CAACGCCG-----ACUUUUCGCACUCAAAUGAGAAUUUUCGUAUAGUGUGUCGAU--UUUGUUCUUGUUUUUUUUUUCUCU ((((((((((((.((.((......))-----.)))))))-----......((((((...(((((....)))))..)))))).....--.)))))))................ ( -21.10, z-score = -0.27, R) >consensus _GAUCGGCGGCGCUUAGUUCUAAGGC___AAUGCCGCCG_____AAGUUUCGCACUUCAAUGAGAAAUUUCAUGUAGUGUGGCGAG__UUCGCAUUGGUUUUUC________ ......(((((((((.......))))......))))).............((((((...(((((....)))))..))))))............................... (-16.76 = -17.16 + 0.40)
| Location | 15,979,611 – 15,979,705 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.31 |
| Shannon entropy | 0.52155 |
| G+C content | 0.50696 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15979611 94 - 21146708 -------CGGAAAACCAAUGCGAA--CUCGCCACACUACAUGAAAUUUCUCAUUGAAGUGCGAAACUU-----CGGCGGCAUU---GCCCCAGAACGAGGCGCCGCCGAUC- -------.((....))...(((..--..)))(.((((.(((((......))).)).)))).).....(-----(((((((...---(((.(.....).)))))))))))..- ( -30.00, z-score = -2.43, R) >droSim1.chr2R 14627287 94 - 19596830 -------GGGAAAAGCAAUGCGAA--CUCGCCACACUACAUGAAAUUUCUCAUUGAAGUGCGAAACUU-----CGGCGGCAUU---GCCCCAGAACGAGGCGCCGCCGAUC- -------............(((..--..)))(.((((.(((((......))).)).)))).).....(-----(((((((...---(((.(.....).)))))))))))..- ( -29.20, z-score = -1.49, R) >droSec1.super_1 13454673 94 - 14215200 -------GGGAAAAGCAAUGCGAA--CUCGCCACACUACAUGAAAUUUCUCACUGAAGUGCGAAACUU-----CGGCGGCAUU---GCCCCAGAACGAGGCGCCGCGUAUC- -------(((....(((((((...--..((((........(((......)))..(((((.....))))-----))))))))))---)))))...(((.(....).)))...- ( -27.70, z-score = -1.01, R) >droYak2.chr2R 7910435 95 + 21139217 -------CGGAAAUCCAAUGCGAACUCUCGCCACACUACAUGAAAUUUCUCAUCGAAGUGCGAAACUU-----CGGCGGCAAU---GCCCCAGAACGAAGCGCCGCCGUC-- -------.((....))...((((....))))(.((((.(((((......)))).).)))).)......-----(((((((...---((..(.....)..)))))))))..-- ( -25.80, z-score = -1.13, R) >droEre2.scaffold_4845 10127598 94 - 22589142 -------CGCCAAUCCAAUGCGAA--CUCGCCACACUACAUGAGAUUUCUCAUUCAAGUGCGAAACUC-----CGGCGGCAAU---GCCCCAGAACGAGGCGCCGCCGAUC- -------............(((..--..)))(.((((..(((((....)))))...)))).)......-----(((((((...---(((.(.....).))))))))))...- ( -30.50, z-score = -2.66, R) >droAna3.scaffold_13266 10523518 96 + 19884421 --------AAGAAUCCGAAACAAA--CUCGCCAUAGUACAUGAAAAUUCUCAUUCAAGUGCGAAACAU-----CGGCGGCAACAAGGCCUUAGAACUAAGCGCCGCCUGUC- --------................--...(((...((((.((((........)))).))))((....)-----))))(((.....(((((((....)))).))))))....- ( -22.10, z-score = -1.31, R) >dp4.chr3 13113887 97 - 19779522 --------AAGAAACCAAAACGAU--CUCGACACACUACAUGAAAUUUCUCAUUGAAGUGCGAAACGCCGCGGCGUCGGCGGCAACGCCUUAGAAGUAAGCGCCGCU----- --------............((..--..)).(.((((.(((((......))).)).)))).).......(((((((.((((....))))(((....)))))))))).----- ( -28.10, z-score = -1.22, R) >droPer1.super_2 526140 97 + 9036312 --------AAGAAACCAAAACGAU--CUCGACACACUACAUGAAAUUUCUCAUUGAAGUGCGAAACGCCGCGGCGUCGGCGGCAACGCCUUAGAAGUAAGCGCCGCU----- --------............((..--..)).(.((((.(((((......))).)).)))).).......(((((((.((((....))))(((....)))))))))).----- ( -28.10, z-score = -1.22, R) >droWil1.scaffold_180701 2354772 88 - 3904529 -------AGAAUAAGGGGGAAAAA--CUCGAAACACUUUAUGAAAUUUCUCAUUGAAGUGCGAAAUGU-----CGGCUAAU-----GCCUUAGAACUAAGCGCCGCC----- -------.......((.((.....--.(((...((((((((((......))).)))))))))).....-----.(((....-----))).............)).))----- ( -18.30, z-score = -0.46, R) >droMoj3.scaffold_6496 21552950 100 - 26866924 AGAGAAAAAAAAAACAAGAACAAA--AUCGACACACUAUACGAAAAUUCUCAUUUGAGUGCGAAAAGU-----CGGCGUUG-----CUGUUAGAAGUAAGCGCCGCUGCUCU ((((....................--.(((.(((.(.....((......))....).))))))..((.-----((((((((-----((......))).))))))))).)))) ( -21.60, z-score = -0.69, R) >consensus ________GAAAAACCAAUACGAA__CUCGCCACACUACAUGAAAUUUCUCAUUGAAGUGCGAAACUU_____CGGCGGCAAU___GCCCCAGAACGAAGCGCCGCCGAUC_ ...........................(((.(((.....((((......))))....))))))...........((((((.....................))))))..... (-12.12 = -12.77 + 0.65)

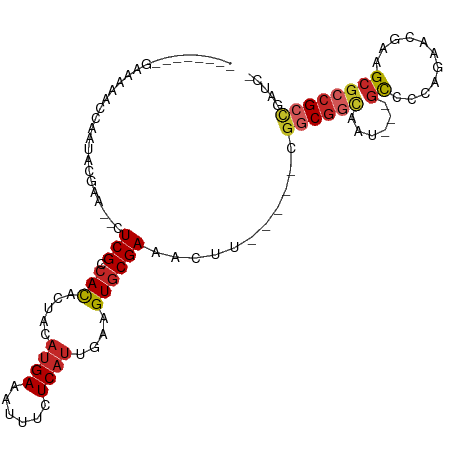
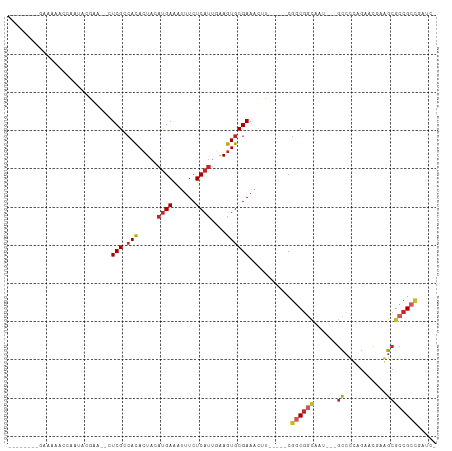
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:16 2011