
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,965,868 – 15,965,976 |
| Length | 108 |
| Max. P | 0.664347 |

| Location | 15,965,868 – 15,965,976 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Shannon entropy | 0.30169 |
| G+C content | 0.39244 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15965868 108 + 21146708 CACCGAC-AACCCACACAGG------GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGC ..(((((-(((((.....))------))................(((((((((.....)))))..))))......))))....(((((((((((.....)))))))))))..)). ( -28.20, z-score = -2.61, R) >droYak2.chr2R 7896427 108 - 21139217 CACCGAC-AACCCACACAGG------GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGC ..(((((-(((((.....))------))................(((((((((.....)))))..))))......))))....(((((((((((.....)))))))))))..)). ( -28.20, z-score = -2.61, R) >droEre2.scaffold_4845 10114162 108 + 22589142 CACCGAC-GACCCACACAGG------GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGGCACUCGUUUGUCAAGCAGCC ....(((-(((((.....))------))................(((((((((.....)))))..))))......))))....(((((((((((.....)))))))))))..... ( -25.20, z-score = -1.16, R) >droSec1.super_1 13441073 108 + 14215200 CACCGAC-AACCCACACAGG------GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGC ..(((((-(((((.....))------))................(((((((((.....)))))..))))......))))....(((((((((((.....)))))))))))..)). ( -28.20, z-score = -2.61, R) >droSim1.chr2R 14611737 108 + 19596830 CACCGAC-AACCCACACAGG------GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGC ..(((((-(((((.....))------))................(((((((((.....)))))..))))......))))....(((((((((((.....)))))))))))..)). ( -28.20, z-score = -2.61, R) >droAna3.scaffold_13266 10510160 106 - 19884421 ------CACUCAGAGUCCGGGGA---GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACGGGC ------........(((((....---...............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....))))))))...))))) ( -24.60, z-score = -1.33, R) >dp4.chr3 13099510 101 + 19779522 --------CUCGA---CUGGAGA---GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACAGGC --------.....---(((....---...............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....))))))))...))).. ( -20.70, z-score = -0.64, R) >droPer1.super_2 511588 101 - 9036312 --------CUCGA---CUGGAGA---GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACAGGC --------.....---(((....---...............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....))))))))...))).. ( -20.70, z-score = -0.64, R) >droWil1.scaffold_180701 2329026 115 + 3904529 CUCAGACAGACGCAGACAGGCUGACAGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAAUUUGAUAAAUGACACUCGUUUGUCAAACGGGC (((.....(((((......((.....))................(((((((((.....)))))..)))).....)))))....(((((((((((.....))))))))))).))). ( -25.60, z-score = -0.95, R) >droVir3.scaffold_12875 17035563 98 - 20611582 ----------CACGGGCAACA------UAUAAUAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACAAC- ----------(((.(....).------.................(((((((((.....)))))..)))).....)))......(((((((((((.....)))))))))))....- ( -20.40, z-score = -1.73, R) >droMoj3.scaffold_6496 21530094 102 + 26866924 ----------CACGGGCAACACAC--AUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACGGA- ----------..(((....)((((--.(((....))).......(((((((((.....)))))..)))).....)))).....(((((((((((.....)))))))))))))..- ( -20.80, z-score = -1.24, R) >droGri2.scaffold_15112 2578750 98 + 5172618 ----------CACGGCCAACA------UAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAAC- ----------...........------..................((((((((.....)))))((((((.....))))))....((((((((((.....)))))))))))))..- ( -19.60, z-score = -0.99, R) >consensus ______C_AACCCACACAGG______GUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACAGGC ............................................(((.......)))......((((((.....))))))...(((((((((((.....)))))))))))..... (-17.53 = -17.28 + -0.25)

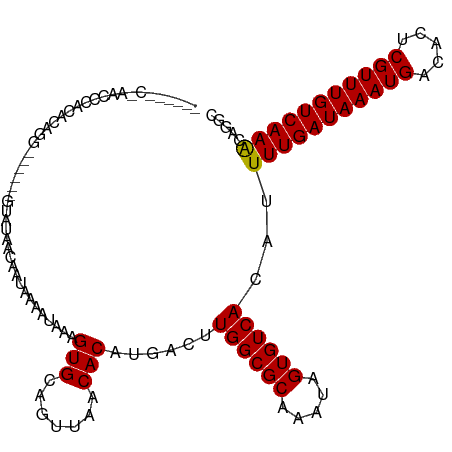
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:13 2011