
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,956,830 – 15,956,898 |
| Length | 68 |
| Max. P | 0.829128 |

| Location | 15,956,830 – 15,956,898 |
|---|---|
| Length | 68 |
| Sequences | 3 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 62.62 |
| Shannon entropy | 0.52746 |
| G+C content | 0.48585 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -8.95 |
| Energy contribution | -8.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
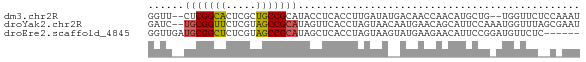
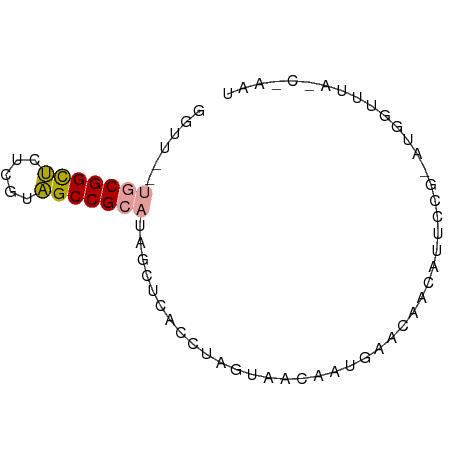
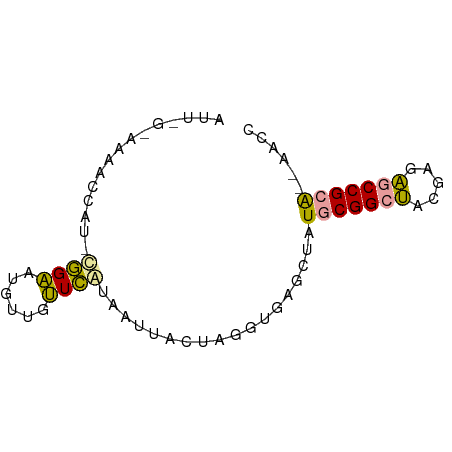
>dm3.chr2R 15956830 68 + 21146708 GGUU--CUCGGCACUCGCUGCCGCAUACCUCACCUUGAUAUGACAACCAACAUGCUG--UGGUUCUCCAAAU (((.--..(((((.....)))))...)))............((.(((((........--))))).))..... ( -15.20, z-score = -1.24, R) >droYak2.chr2R 20933168 70 - 21139217 GAUC--UGCGGUUCUCGUAGCCGCAUAGUUCACCUAGUAACAAUGAACAGCAUUCCAAAUGGUUUAGCGAAU ....--(((((((.....)))))))..(((((...........))))).(((..((....))..).)).... ( -15.00, z-score = -0.99, R) >droEre2.scaffold_4845 20419854 66 - 22589142 GGUUGAUGCGGCUCUCGUAGCCGCAUAGCUCACCUAGUAAGUAUGAAGAACAUUCCGGAUGUUCUC------ (((..((((((((.....)))))))).....)))............((((((((...)))))))).------ ( -20.70, z-score = -1.97, R) >consensus GGUU__UGCGGCUCUCGUAGCCGCAUAGCUCACCUAGUAACAAUGAACAACAUUCCG_AUGGUUUA_C_AAU ((....(((((((.....)))))))....))......................................... ( -8.95 = -8.73 + -0.22)
| Location | 15,956,830 – 15,956,898 |
|---|---|
| Length | 68 |
| Sequences | 3 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 62.62 |
| Shannon entropy | 0.52746 |
| G+C content | 0.48585 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -9.39 |
| Energy contribution | -8.40 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
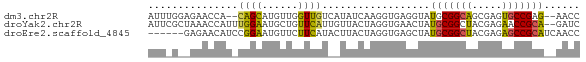
>dm3.chr2R 15956830 68 - 21146708 AUUUGGAGAACCA--CAGCAUGUUGGUUGUCAUAUCAAGGUGAGGUAUGCGGCAGCGAGUGCCGAG--AACC .....((.(((((--........))))).))............(((...(((((.....)))))..--.))) ( -17.50, z-score = -0.46, R) >droYak2.chr2R 20933168 70 + 21139217 AUUCGCUAAACCAUUUGGAAUGCUGUUCAUUGUUACUAGGUGAACUAUGCGGCUACGAGAACCGCA--GAUC ....((....((....))...)).(((((((.......)))))))..(((((.........)))))--.... ( -14.50, z-score = -0.39, R) >droEre2.scaffold_4845 20419854 66 + 22589142 ------GAGAACAUCCGGAAUGUUCUUCAUACUUACUAGGUGAGCUAUGCGGCUACGAGAGCCGCAUCAACC ------((((((((.....))))))))...........(((.....((((((((.....))))))))..))) ( -20.80, z-score = -2.64, R) >consensus AUU_G_AAAACCAU_CGGAAUGUUGUUCAUAAUUACUAGGUGAGCUAUGCGGCUACGAGAGCCGCA__AACC ...............((((......)))).........(((......(((((((.....)))))))...))) ( -9.39 = -8.40 + -0.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:12 2011