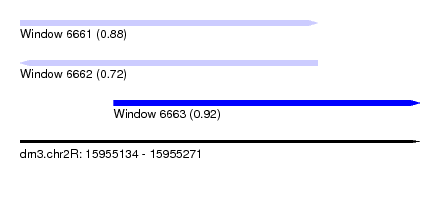
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,955,134 – 15,955,271 |
| Length | 137 |
| Max. P | 0.923311 |

| Location | 15,955,134 – 15,955,236 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.64 |
| Shannon entropy | 0.40467 |
| G+C content | 0.50444 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
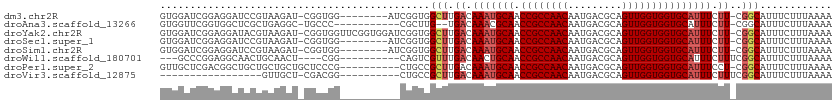
>dm3.chr2R 15955134 102 + 21146708 GUGGAUCGGAGGAUCCGUAAGAU-CGGUGG--------AUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUUCUUUAAAA ..(((((....)))))....(((-(....)--------)))((..(((.((.(((((((.((((((((.........))))))))))))))).))-.)))...))....... ( -35.60, z-score = -2.23, R) >droAna3.scaffold_13266 10500941 97 - 19884421 GUGGUUCGGUGGCUCGCUGAGGC-UGCCC-----------CGCUUG--UGACAAACGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUUCUUUAAAA ((((...((..(((......)))-..)))-----------)))...--........(((.((((((((.........))))))))))).......-................ ( -29.90, z-score = 0.12, R) >droYak2.chr2R 7887893 110 - 21139217 GUGGAUCGGAGGAUACGUAAGAU-CGGUGGUUCGGUGGAUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUUCUUUAAAA ...(.((((((.....(((((.(-((.(((((.....))))).)))))).))(((((((.((((((((.........))))))))))))))))))-))))............ ( -34.00, z-score = -1.15, R) >droSec1.super_1 13432880 102 + 14215200 GUGGAUCGGAGGAUCCGUAAGAU-CGGUGG--------AUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUUCUUUAAAA ..(((((....)))))....(((-(....)--------)))((..(((.((.(((((((.((((((((.........))))))))))))))).))-.)))...))....... ( -35.60, z-score = -2.23, R) >droSim1.chr2R 14603539 102 + 19596830 GUGGAUCGGAGGAUCCGUAAGAU-CGGUGG--------AUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUUCUUUAAAA ..(((((....)))))....(((-(....)--------)))((..(((.((.(((((((.((((((((.........))))))))))))))).))-.)))...))....... ( -35.60, z-score = -2.23, R) >droWil1.scaffold_180701 2314000 95 + 3904529 ---GCCCGGAGGCAACUGCAACU----CGG----------CAGUCGUUUGACAACUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCUUUAAAA ---(((.(((((.((.(((....----..(----------((((.(.....).))))).(((((((((.........)))))))))))).))))))))))............ ( -33.10, z-score = -2.37, R) >droPer1.super_2 503006 101 - 9036312 GUUGCUCGACGGCUGCUGCUGCUGCUCCCG----------CUGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCCU-CGGCAUUUCUUUAAAA ..(((.(((((((.((.((....))....)----------).))))......(((((((.((((((((.........)))))))))))))))..)-)))))........... ( -35.40, z-score = -2.07, R) >droVir3.scaffold_12875 17016504 84 - 20611582 -----------------GUUGCU-CGACGG----------CUGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCUUUAAAA -----------------...(((-....))----------)(((((...((.(((((((.((((((((.........))))))))))))))).)).)))))........... ( -29.20, z-score = -2.58, R) >consensus GUGGAUCGGAGGAUCCGUAAGAU_CGGCGG__________CGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU_CGGCAUUUCUUUAAAA .............................................(((....(((((((.((((((((.........))))))))))))))).....)))............ (-20.63 = -21.04 + 0.41)

| Location | 15,955,134 – 15,955,236 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Shannon entropy | 0.40467 |
| G+C content | 0.50444 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15955134 102 - 21146708 UUUUAAAGAAAUGCCG-AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAU--------CCACCG-AUCUUACGGAUCCUCCGAUCCAC ...............(-(..(((((((((.(((((.........))))).)).))))))).))........(((--------(....)-)))....(((((....))))).. ( -28.00, z-score = -2.39, R) >droAna3.scaffold_13266 10500941 97 + 19884421 UUUUAAAGAAAUGCCG-AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCGUUUGUCA--CAAGCG-----------GGGCA-GCCUCAGCGAGCCACCGAACCAC ...............(-(..(((((((((.(((((.........))))).)).))))))).)).--....((-----------((((.-((....))..))).)))...... ( -29.40, z-score = -0.64, R) >droYak2.chr2R 7887893 110 + 21139217 UUUUAAAGAAAUGCCG-AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAUCCACCGAACCACCG-AUCUUACGUAUCCUCCGAUCCAC .......((.((((.(-(..(((((((((.(((((.........))))).)).))))))).))........((((............)-)))....))))..))........ ( -21.70, z-score = -0.63, R) >droSec1.super_1 13432880 102 - 14215200 UUUUAAAGAAAUGCCG-AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAU--------CCACCG-AUCUUACGGAUCCUCCGAUCCAC ...............(-(..(((((((((.(((((.........))))).)).))))))).))........(((--------(....)-)))....(((((....))))).. ( -28.00, z-score = -2.39, R) >droSim1.chr2R 14603539 102 - 19596830 UUUUAAAGAAAUGCCG-AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAU--------CCACCG-AUCUUACGGAUCCUCCGAUCCAC ...............(-(..(((((((((.(((((.........))))).)).))))))).))........(((--------(....)-)))....(((((....))))).. ( -28.00, z-score = -2.39, R) >droWil1.scaffold_180701 2314000 95 - 3904529 UUUUAAAGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAGUUGUCAAACGACUG----------CCG----AGUUGCAGUUGCCUCCGGGC--- .............(((...((..((((((.(((((.........))))).)).))))....))...((((((----------(..----....)))))))....)))..--- ( -28.70, z-score = -0.77, R) >droPer1.super_2 503006 101 + 9036312 UUUUAAAGAAAUGCCG-AGGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGCAG----------CGGGAGCAGCAGCAGCAGCCGUCGAGCAAC ...........(((((-((((((((((((.(((((.........))))).)).))))))).))...((((.(----------(....)).((....)).))))))).))).. ( -35.70, z-score = -1.40, R) >droVir3.scaffold_12875 17016504 84 + 20611582 UUUUAAAGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGCAG----------CCGUCG-AGCAAC----------------- .......((..(((((..(.(((((((((.(((((.........))))).)).))))))).)....))))).----------...)).-......----------------- ( -27.80, z-score = -2.10, R) >consensus UUUUAAAGAAAUGCCG_AAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCG__________CCACCG_AUCUUACGGAUCCUCCGAUCCAC ....................(((((((((.(((((.........))))).)).))))))).................................................... (-16.98 = -17.00 + 0.02)

| Location | 15,955,166 – 15,955,271 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Shannon entropy | 0.37395 |
| G+C content | 0.45071 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15955166 105 + 21146708 -----GGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUCGCUUUGCUGUGCC---- -----((((((((((.(((((((.((((((((.........)))))))))))))))..)-))((((..-.((((....))))...))))))))))-)..(((......)))..---- ( -35.10, z-score = -2.34, R) >droAna3.scaffold_13266 10500970 107 - 19884421 -------GCUUGUGACAAACGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGCCCGC-CGCCGCCUUCGCUGGCGAGUC -------((..(((......(((.((((((((.........))))))))))).......-.(((((..-...((((......)))).))))))))-.))((((......)))).... ( -31.80, z-score = -1.00, R) >droEre2.scaffold_4845 10105998 105 + 22589142 -----GGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUCGCUUGGCUGGGCC---- -----((((((((((.(((((((.((((((((.........)))))))))))))))..)-))((((..-.((((....))))...))))))))))-)...((((....)))).---- ( -36.30, z-score = -2.01, R) >droYak2.chr2R 7887933 105 - 21139217 -----GGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUCGCAUGGCUGGGCC---- -----((((((((((.(((((((.((((((((.........)))))))))))))))..)-))((((..-.((((....))))...))))))))))-).......(((...)))---- ( -35.10, z-score = -1.51, R) >droSec1.super_1 13432912 105 + 14215200 -----GGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUCGCUUUGCUGGGCC---- -----((((((((((.(((((((.((((((((.........)))))))))))))))..)-))((((..-.((((....))))...))))))))))-)...((((....)))).---- ( -36.40, z-score = -2.38, R) >droSim1.chr2R 14603571 105 + 19596830 -----GGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUCGCUUUGCUGGGCC---- -----((((((((((.(((((((.((((((((.........)))))))))))))))..)-))((((..-.((((....))))...))))))))))-)...((((....)))).---- ( -36.40, z-score = -2.38, R) >droWil1.scaffold_180701 2314024 98 + 3904529 -----AGUCGUUUGACAACUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUU-CUUUAAAAAUAAAUUUAUGUGGCCGC-CUUUGUCUC------------ -----........(((((.((((.((((((((.........)))))))))))).......((((....-...((((......))))....)))).-..)))))..------------ ( -26.00, z-score = -1.70, R) >droPer1.super_2 503037 87 - 9036312 -----UGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCCU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUG--UGC-C-------------------- -----(((((...((..((((((.((((((((.........))))))))))))))))..-)))))...-.....................--...-.-------------------- ( -26.40, z-score = -2.33, R) >dp4.chr3 13090934 87 + 19779522 -----UGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCCU-CGGCAUUU-CUUUAAAAAUAAAUUUAUGUG--CGC-C-------------------- -----(((((...((..((((((.((((((((.........))))))))))))))))..-)))))...-.....................--...-.-------------------- ( -26.40, z-score = -2.14, R) >droVir3.scaffold_12875 17016520 100 - 20611582 --------CGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUU-CUUUAAAAAUAAAUUUAUGUGAUAAUUCCUCGUAUUGGCC-------- --------.(((..(((((((((.((((((((.........))))))))))))))).....((.((((-(..((((......))))...)).))).))..))...))).-------- ( -25.50, z-score = -1.78, R) >droMoj3.scaffold_6496 21517438 101 + 26866924 --------CGCUUGACAAAUGCAACCGCCAACAAAGACGCAGUUGGUGGUGCAUUUCUUUGGGCAUUUCUUUUAAAAAUAAAUUUAUGUGAUAAUUCCUCGUAUUGGCG-------- --------.(((..(.(((((((.((((((((.........)))))))))))))))..)..)))....................((((.((....))..))))......-------- ( -27.50, z-score = -2.14, R) >droGri2.scaffold_15112 2568208 109 + 5172618 UGCUGCCGCAUUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUUCUUUAAAAAUAAAUUUAUGUGAUAAUUCCUCGUAUUGGCC-------- .(((((((.....((.(((((((.((((((((.........))))))))))))))).)).))))....................((((.((....))..))))..))).-------- ( -29.30, z-score = -1.81, R) >consensus _____GGUCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUU_CGGCAUUU_CUUUAAAAAUAAAUUUAUGUGGCCGC_CUUCGCAUUGCUG________ .........(((....(((((((.((((((((.........))))))))))))))).....)))..................................................... (-20.60 = -20.82 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:11 2011