
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,951,363 – 15,951,454 |
| Length | 91 |
| Max. P | 0.912059 |

| Location | 15,951,363 – 15,951,454 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Shannon entropy | 0.40211 |
| G+C content | 0.43039 |
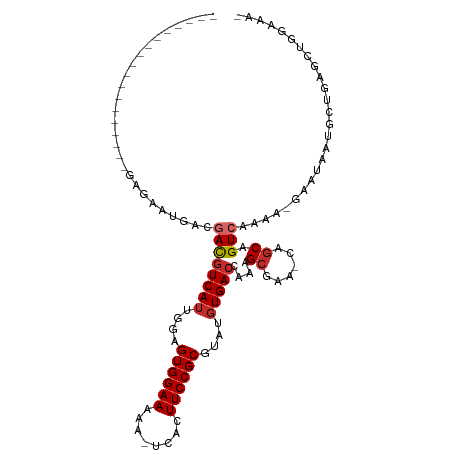
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
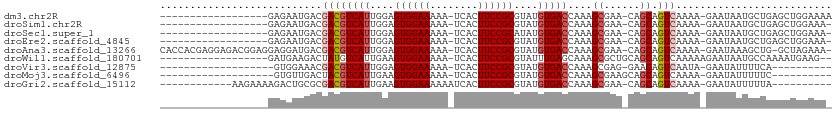
>dm3.chr2R 15951363 91 + 21146708 ------------------GAGAAUGACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAAUGCUGAGCUGGAAAA ------------------............(((((....((((((...-....))))))....)))))...(((...-(((((.((....-))....))))).)))...... ( -23.00, z-score = -1.84, R) >droSim1.chr2R 14599776 90 + 19596830 ------------------GAGAAUGACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAAUGCUGAGCUGGAAA- ------------------............(((((....((((((...-....))))))....)))))...(((...-(((((.((....-))....))))).))).....- ( -23.00, z-score = -1.79, R) >droSec1.super_1 13429170 90 + 14215200 ------------------GAGAAUGACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCAUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAAUGCUGAGCUGGAAA- ------------------............(((((....((((((...-....))))))....)))))...(((...-(((((.((....-))....))))).))).....- ( -21.90, z-score = -1.71, R) >droEre2.scaffold_4845 10102204 90 + 22589142 ------------------GAGAAUGACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAAUGCUGAGCUGGAAA- ------------------............(((((....((((((...-....))))))....)))))...(((...-(((((.((....-))....))))).))).....- ( -23.00, z-score = -1.79, R) >droAna3.scaffold_13266 10497430 107 - 19884421 CACCACGAGGAGACGGAGGAGGAUGACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAAAGCUG-GCUAGAAA- (((.(((.(((((((..(........)..)))).....(((((.....-)))))))).)))..))).....(((...-((((..((....-)).....))))-))).....- ( -23.60, z-score = -0.29, R) >droWil1.scaffold_180701 2309771 91 + 3904529 ------------------GAUGAAGACUAUGUCAUUGAAGUGGAAAAA-UCACUUCCGCGUAUUUGAGCAAAGCGCUGCAGCAGUCAAAAAGAAUAAUGCCAAAAUGAAG-- ------------------......((((.(((....(((((((.....-))))))).((((.(((....))))))).)))..))))........................-- ( -16.50, z-score = -0.06, R) >droVir3.scaffold_12875 17012647 80 - 20611582 -------------------GUGGAAACGACGUCAUUGGAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAG-GAACAGUCAAUA-GAAUAUUUUCA---------- -------------------(((....).))(((((....((((((...-....))))))....)))))......(((-((....((....-))...))))).---------- ( -17.20, z-score = -0.42, R) >droMoj3.scaffold_6496 21512723 81 + 26866924 -------------------GUGUUGACUACGUCAUUGAAGUGGAAAAA-UCACUUCCGCGUAUGUGACCAAAGCGAAGCAGCAGUCAAAA-GAAUAUUUUUC---------- -------------------...((((((..(((((....((((((...-....))))))....)))))....((......))))))))..-...........---------- ( -21.30, z-score = -2.33, R) >droGri2.scaffold_15112 2563627 88 + 5172618 ------------AAGAAAAGACUGCGCGACGUCAUUGAAGUGGAAAAAAUCACUUCCGCGUAUGUGACCAAAGCGAA-CAGCAGUCAAAA-GAAUAUUUUUA---------- ------------.......((((((.((..(((((....((((((........))))))....))))).....))..-..))))))....-...........---------- ( -25.10, z-score = -3.33, R) >consensus __________________GAGAAUGACGACGUCAUUGGAGUGGAAAAA_UCACUUCCGCGUAUGUGACCAAAGCGAA_CAGCAGUCAAAA_GAAUAAUGCUGAGCUGGAAA_ ...........................((((((((....((((((........))))))....)))))....((......)).))).......................... (-16.26 = -16.72 + 0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:09 2011