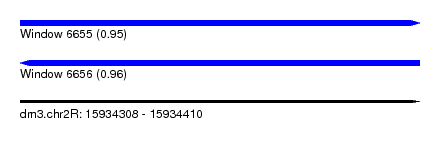
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,934,308 – 15,934,410 |
| Length | 102 |
| Max. P | 0.958490 |

| Location | 15,934,308 – 15,934,410 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.05 |
| Shannon entropy | 0.33827 |
| G+C content | 0.49237 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
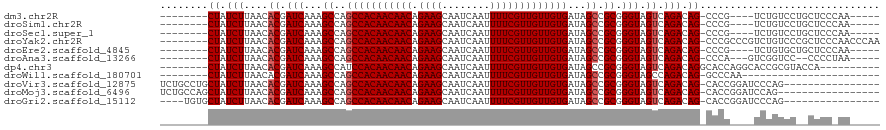
>dm3.chr2R 15934308 102 + 21146708 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCG----UCUGUCCUGCUCCCAA----- --------...................((..(((((((((((.((((........)))))))))))))...)).))(((.(((..(((((-....----.)))))..))))))..----- ( -29.00, z-score = -3.14, R) >droSim1.chr2R 14582789 102 + 19596830 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCG----UCUGUCCUGCUCCCAA----- --------...................((..(((((((((((.((((........)))))))))))))...)).))(((.(((..(((((-....----.)))))..))))))..----- ( -29.00, z-score = -3.14, R) >droSec1.super_1 13412177 102 + 14215200 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCG----UCUGUCCUGCUCCCAA----- --------...................((..(((((((((((.((((........)))))))))))))...)).))(((.(((..(((((-....----.)))))..))))))..----- ( -29.00, z-score = -3.14, R) >droYak2.chr2R 7863199 111 - 21139217 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCGCCCGUCUGUCCCGCUCCCAACCCAA --------...................((.(..(((((((((.((((........))))))))))))).).)).(((((....(((((..-.......))))).)))))........... ( -29.60, z-score = -3.43, R) >droEre2.scaffold_4845 10085330 102 + 22589142 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCG----UCUGUGCUGCUCCCAA----- --------...................((..(((((((((((.((((........)))))))))))))...)).))((((((((((((..-...)----)))).)))))))....----- ( -30.30, z-score = -2.85, R) >droAna3.scaffold_13266 10480902 101 - 19884421 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CCCA---GUCGGUCC--CCCCUAA----- --------............((((...((..(((((((((((.((((........)))))))))))))...)).))((((.(.....).)-))).---...)))).--.......----- ( -25.20, z-score = -2.53, R) >dp4.chr3 13061875 102 + 19779522 --------CUAUCUUAACACGAUCAAAGCCAUCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAGGCACCAGGCACCGCGUACCA---------- --------........((.((............(((((((((.((((........)))))))))))))...(((...(((.(((.....)))))).)))..)).))....---------- ( -27.30, z-score = -2.49, R) >droWil1.scaffold_180701 2268277 88 + 3904529 --------CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGCCAGACAG-GCCCAA----------------------- --------...................(((.(((((((((((.((((........)))))))))))))......)).))).(((.....)-))....----------------------- ( -20.30, z-score = -1.47, R) >droVir3.scaffold_12875 16985084 103 - 20611582 UCUGCCUGCUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CACCGGAUCCCAG---------------- .((((((((.(((.......)))....((.(..(((((((((.((((........))))))))))))).).)).))))))))........-.............---------------- ( -25.90, z-score = -1.57, R) >droMoj3.scaffold_6496 21484346 102 + 26866924 UCUGCCAGCUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CACCGGAUCCAG----------------- ((((...(((.(((....((.(((...((..(((((((((((.((((........)))))))))))))...)).)).))).)).))).))-)..)))).....----------------- ( -24.00, z-score = -1.24, R) >droGri2.scaffold_15112 2543361 99 + 5172618 ----UGUGCUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG-CACCGGAUCCCAG---------------- ----.(((((.(((....((.(((...((..(((((((((((.((((........)))))))))))))...)).)).))).)).))).))-)))..........---------------- ( -25.40, z-score = -1.83, R) >consensus ________CUAUCUUAACACGAUCAAAGCCAGCCACAACAACAGAAGCAAUCAAUUUUCGUUGUUGUGAUAGCCGCGGGUAGUCAGACAG_CCCC____UCUGUCC__CUCC_AA_____ ....................(((....(((.(((((((((((.((((........)))))))))))))......)).))).))).................................... (-16.40 = -16.58 + 0.18)

| Location | 15,934,308 – 15,934,410 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.05 |
| Shannon entropy | 0.33827 |
| G+C content | 0.49237 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -25.67 |
| Energy contribution | -25.59 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
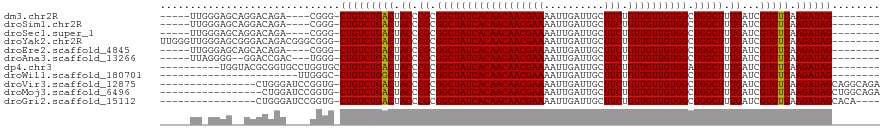
>dm3.chr2R 15934308 102 - 21146708 -----UUGGGAGCAGGACAGA----CGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----..(((((..((((((.----....-))))))..)).)))..((((((((((((((((((..........))).)))))))))).)))))...(((.......)))..-------- ( -33.40, z-score = -2.06, R) >droSim1.chr2R 14582789 102 - 19596830 -----UUGGGAGCAGGACAGA----CGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----..(((((..((((((.----....-))))))..)).)))..((((((((((((((((((..........))).)))))))))).)))))...(((.......)))..-------- ( -33.40, z-score = -2.06, R) >droSec1.super_1 13412177 102 - 14215200 -----UUGGGAGCAGGACAGA----CGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----..(((((..((((((.----....-))))))..)).)))..((((((((((((((((((..........))).)))))))))).)))))...(((.......)))..-------- ( -33.40, z-score = -2.06, R) >droYak2.chr2R 7863199 111 + 21139217 UUGGGUUGGGAGCGGGACAGACGGGCGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- ...(((..(..(((((.(((((((.....-)))))))....)))))((((((((((((((((((..........))).)))))))))).))))))..)))............-------- ( -40.70, z-score = -2.76, R) >droEre2.scaffold_4845 10085330 102 - 22589142 -----UUGGGAGCAGCACAGA----CGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----..((((((((.((((.----....-))))))).)).)))..((((((((((((((((((..........))).)))))))))).)))))...(((.......)))..-------- ( -32.10, z-score = -1.48, R) >droAna3.scaffold_13266 10480902 101 + 19884421 -----UUAGGGG--GGACCGAC---UGGG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----....((.--...))...---....-(((((((((.((.((.((((((((((((((((((..........))).)))))))))).))))).))...))))).))))))-------- ( -30.50, z-score = -1.39, R) >dp4.chr3 13061875 102 - 19779522 ----------UGGUACGCGGUGCCUGGUGCCUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGAUGGCUUUGAUCGUGUUAAGAUAG-------- ----------.(((((.(((...))))))))((((((((.((.((.((((((((((((((((((..........))).)))))))).))))))).))...))))).))))).-------- ( -31.90, z-score = -1.84, R) >droWil1.scaffold_180701 2268277 88 - 3904529 -----------------------UUGGGC-CUGUCUGGCUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG-------- -----------------------...(((-(.....))))...((.((((((((((((((((((..........))).)))))))))).))))).))(((.......)))..-------- ( -25.60, z-score = -1.48, R) >droVir3.scaffold_12875 16985084 103 + 20611582 ----------------CUGGGAUCCGGUG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAGCAGGCAGA ----------------((((...))))((-(((((((((.((.((.((((((((((((((((((..........))).)))))))))).))))).))...))))).))))))))...... ( -33.20, z-score = -1.48, R) >droMoj3.scaffold_6496 21484346 102 - 26866924 -----------------CUGGAUCCGGUG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAGCUGGCAGA -----------------(((...(((..(-(((((((((.((.((.((((((((((((((((((..........))).)))))))))).))))).))...))))).))))))))))))). ( -32.40, z-score = -1.83, R) >droGri2.scaffold_15112 2543361 99 - 5172618 ----------------CUGGGAUCCGGUG-CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAGCACA---- ----------------..........(((-(((((((((.((.((.((((((((((((((((((..........))).)))))))))).))))).))...))))).))))))))).---- ( -34.90, z-score = -3.16, R) >consensus _____UU_GGAG__GGACAGA____CGGG_CUGUCUGACUACCCGCGGCUAUCACAACAACGAAAAUUGAUUGCUUCUGUUGUUGUGGCUGGCUUUGAUCGUGUUAAGAUAG________ ..............................(((((((((.((.((.((((((((((((((((((..........))).)))))))))).))))).))...))))).))))))........ (-25.67 = -25.59 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:05 2011