| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,928,302 – 15,928,433 |
| Length | 131 |
| Max. P | 0.998130 |

| Location | 15,928,302 – 15,928,402 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 62.48 |
| Shannon entropy | 0.72453 |
| G+C content | 0.44648 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -9.78 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
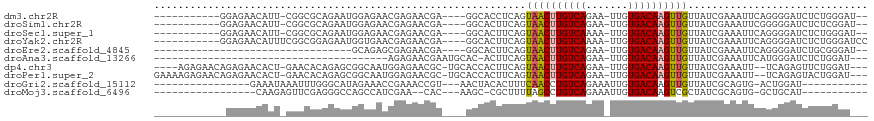
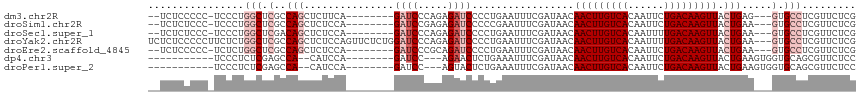
>dm3.chr2R 15928302 100 + 21146708 -----------GGAGAACAUU-CGGCGCAGAAUGGAGAACGAGAACGA----GGCACCUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUCUGGGAU-- -----------(((((.((((-(......)))))(((..(((.(((((----(....)))...(((((((((...-...)))))))))))).)))...)))......))))).....-- ( -29.20, z-score = -1.80, R) >droSim1.chr2R 14576788 100 + 19596830 -----------GGAGAACAUU-CGGCGCAGAAUGGAGAACGAGAACGA----GGCACUUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUUCGGGGGAUCUCUCGGAU-- -----------......((((-(......))))).....(((.(((((----(....)))...(((((((((...-...)))))))))))).)))..((((((((....))))))))-- ( -27.30, z-score = -1.10, R) >droSec1.super_1 13406170 100 + 14215200 -----------GGAGAACAUU-CGGCGCAGAAUGGAGAACGAGAACGA----GGCACUUCAGUAACUUGUCAAAA-UUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUCUGGGAU-- -----------(((((.((((-(......)))))(((..(((.(((((----(....)))...(((((((((...-...)))))))))))).)))...)))......))))).....-- ( -26.00, z-score = -1.33, R) >droYak2.chr2R 7857255 103 - 21139217 -----------GGAGAACAUUUCGGCGGAGAAUGGUGAACGAGAACGA----GGCACUUCAGUAACUUGUCAAAA-UUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUCUGGGAUCC -----------...((..(((((((..(.((..((((..((....)).----..))))))..((((((((((...-...)))))))))))..)))))))))...(((((.....))))) ( -26.00, z-score = -0.49, R) >droEre2.scaffold_4845 10079544 79 + 22589142 ---------------------------------GCAGAGCGAGAACGA----GGCACUUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUGCGGGAU-- ---------------------------------(((((.(((.(((((----(....)))...(((((((((...-...)))))))))))).)))....((....))))))).....-- ( -21.40, z-score = -1.41, R) >droAna3.scaffold_13266 10474799 75 - 19884421 ---------------------------------------AGAGAACGAAUGCAC-ACUUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUUCAUGGGAUCUCUGGAU--- ---------------------------------------(((((.(..(((...-.......((((((((((...-...))))))))))...........)))..).)))))....--- ( -16.80, z-score = -0.46, R) >dp4.chr3 13054803 107 + 19779522 ----AGAGAACAGAGAACACU-GAACACAGAGCGGCAAUGGAGAACGC-UGCACCACUUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUU--UCAGAGUUCUGGAU--- ----.........(((((.((-(((....(((((((..........))-)))..........((((((((((...-...))))))))))...)).....)--)))).)))))....--- ( -29.40, z-score = -1.63, R) >droPer1.super_2 473048 111 - 9036312 GAAAAGAGAACAGAGAACACU-GAACACAGAGCGGCAAUGGAGAACGC-UGCACCACUUCAGUAACUUGUCAGAA-UUGUGACAAGUUGUUAUCGAAAUU--UCAGAGUACUGGAU--- ............((.((((((-(((....(.(((((..........))-))).)...))))))(((((((((...-...)))))))))))).)).....(--((((....))))).--- ( -27.40, z-score = -1.42, R) >droGri2.scaffold_15112 2535226 88 + 5172618 ----------------GAAAUAAAUUUGGGCAUAGAAACCGAAACCGU---AACUACACUUUCAACCUGUCAGAAAUUGUGACAAGUUGUUAUCGCAGUG-ACUGGAU----------- ----------------........(((((.........))))).((((---.(((.......((((.(((((.......))))).)))).......))).-)).))..----------- ( -16.14, z-score = -0.32, R) >droMoj3.scaffold_6496 21474527 84 + 26866924 -----------------CAAGAGUUCGAGGGCCAGCCAUCGAA--CAC---AAGC-CGCUUUUAGCCUGUCAGAAAUUGUGACAAGUCGCUAUCGCAGUG-GCUGCAU----------- -----------------.....((((((.((....)).)))))--)..---.(((-((((..((((.(((((.......)))))....))))....))))-)))....----------- ( -28.70, z-score = -2.16, R) >consensus ____________GAGAACAUU_CGGCGCAGAAUGGAGAACGAGAACGA____AGCACUUCAGUAACUUGUCAGAA_UUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUCUGGGA___ ..............................................................((((((((((.......)))))))))).............................. ( -9.78 = -9.90 + 0.12)

| Location | 15,928,302 – 15,928,402 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 62.48 |
| Shannon entropy | 0.72453 |
| G+C content | 0.44648 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -9.48 |
| Energy contribution | -9.33 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15928302 100 - 21146708 --AUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAGGUGCC----UCGUUCUCGUUCUCCAUUCUGCGCCG-AAUGUUCUCC----------- --......((((......(((...(((.((((((((((((...-...)))))))))...(((....)----))))).))))))..(((((......)-)))).)))).----------- ( -25.10, z-score = -2.90, R) >droSim1.chr2R 14576788 100 - 19596830 --AUCCGAGAGAUCCCCCGAAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAAGUGCC----UCGUUCUCGUUCUCCAUUCUGCGCCG-AAUGUUCUCC----------- --....(((((.......(((...(((.((((((((((((...-...)))))))))...((......----))))).))))))..(((((......)-))))))))).----------- ( -22.00, z-score = -2.01, R) >droSec1.super_1 13406170 100 - 14215200 --AUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAA-UUUUGACAAGUUACUGAAGUGCC----UCGUUCUCGUUCUCCAUUCUGCGCCG-AAUGUUCUCC----------- --......((((......(((...(((.((((((((((((...-...)))))))))...((......----))))).))))))..(((((......)-)))).)))).----------- ( -20.80, z-score = -2.14, R) >droYak2.chr2R 7857255 103 + 21139217 GGAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAA-UUUUGACAAGUUACUGAAGUGCC----UCGUUCUCGUUCACCAUUCUCCGCCGAAAUGUUCUCC----------- (((((.....))))).....((((((.....(((((((((...-...)))))))))...((((((..----.((....))..)))..))).....)))))).......----------- ( -22.80, z-score = -2.07, R) >droEre2.scaffold_4845 10079544 79 - 22589142 --AUCCCGCAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAAGUGCC----UCGUUCUCGCUCUGC--------------------------------- --.....(((((((....))....(((.((((((((((((...-...)))))))))...((......----))))).))).)))))--------------------------------- ( -17.60, z-score = -2.59, R) >droAna3.scaffold_13266 10474799 75 + 19884421 ---AUCCAGAGAUCCCAUGAAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAAGU-GUGCAUUCGUUCUCU--------------------------------------- ---....(((((....(((.((((((.....(((((((((...-...)))))))))..))))))-...)))....)))))--------------------------------------- ( -19.10, z-score = -2.64, R) >dp4.chr3 13054803 107 - 19779522 ---AUCCAGAACUCUGA--AAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAAGUGGUGCA-GCGUUCUCCAUUGCCGCUCUGUGUUC-AGUGUUCUCUGUUCUCU---- ---....(((((...((--(...........(((((((((...-...)))))))))(((((((..(.((.-(((........))).)).)..).)))-))).)))...)))))..---- ( -29.90, z-score = -2.83, R) >droPer1.super_2 473048 111 + 9036312 ---AUCCAGUACUCUGA--AAUUUCGAUAACAACUUGUCACAA-UUCUGACAAGUUACUGAAGUGGUGCA-GCGUUCUCCAUUGCCGCUCUGUGUUC-AGUGUUCUCUGUUCUCUUUUC ---...(((....))).--......((.((((((((((((...-...))))))))((((((((..(.((.-(((........))).)).)..).)))-)))).....)))).))..... ( -27.90, z-score = -2.46, R) >droGri2.scaffold_15112 2535226 88 - 5172618 -----------AUCCAGU-CACUGCGAUAACAACUUGUCACAAUUUCUGACAGGUUGAAAGUGUAGUU---ACGGUUUCGGUUUCUAUGCCCAAAUUUAUUUC---------------- -----------.....((-.((((((....((((((((((.......))))))))))....)))))).---))(((((.(((......))).)))))......---------------- ( -24.40, z-score = -3.31, R) >droMoj3.scaffold_6496 21474527 84 - 26866924 -----------AUGCAGC-CACUGCGAUAGCGACUUGUCACAAUUUCUGACAGGCUAAAAGCG-GCUU---GUG--UUCGAUGGCUGGCCCUCGAACUCUUG----------------- -----------..(((((-(.((((....))(.(((((((.......))))))))....)).)-)).)---))(--(((((.((....)).)))))).....----------------- ( -27.50, z-score = -1.75, R) >consensus ___UCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAA_UUCUGACAAGUUACUGAAGUGCC____UCGUUCUCGUUCUCCAUUCUGCGCCG_AAUGUUCUC____________ ...............................(((((((((.......)))))))))............................................................... ( -9.48 = -9.33 + -0.15)

| Location | 15,928,329 – 15,928,433 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.33264 |
| G+C content | 0.49409 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -12.31 |
| Energy contribution | -13.31 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
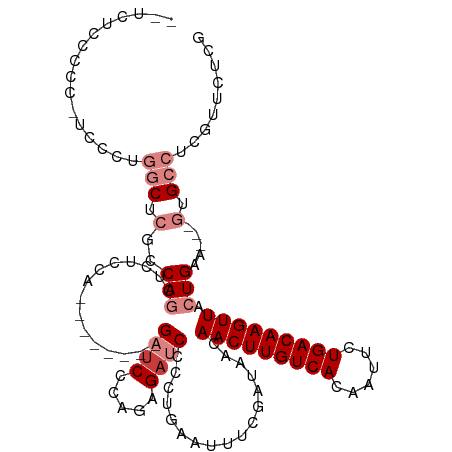
>dm3.chr2R 15928329 104 - 21146708 --UCUCCCCC-UCCCUGGCUCGCCAGCUCUUCA--------GAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAG---GUGCCUCGUUCUCG --........-...((((....))))...((((--------((((.....))))...))))...(((.((((((((((((......)))))))))...(((---....)))))).))) ( -25.60, z-score = -2.40, R) >droSim1.chr2R 14576815 104 - 19596830 --UCUCUCCC-UCCCUGGCUCGCCAGCUCUCCA--------GAUCCGAGAGAUCCCCCGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAA---GUGCCUCGUUCUCG --((((((..-...((((....)))).((....--------))...))))))............(((.((((((((((((......)))))))))...((.---.....))))).))) ( -24.80, z-score = -2.25, R) >droSec1.super_1 13406197 104 - 14215200 --UCUCUCCC-UCCCUGGCUCGACAGCUCUCCA--------GAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAA---GUGCCUCGUUCUCG --........-.....(((.(..(((.((....--------((((.....))))....))...........(((((((((......))))))))).)))..---).)))......... ( -21.60, z-score = -1.44, R) >droYak2.chr2R 7857283 115 + 21139217 UCUCUCCCCCUUCUCUGGCUCGCCAGCUCUCCAGUUCUCUGGAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAA---GUGCCUCGUUCUCG ................(((.(..(((.....(((.(((((((...)))))))....)))............(((((((((......))))))))).)))..---).)))......... ( -29.20, z-score = -3.02, R) >droEre2.scaffold_4845 10079550 104 - 22589142 --UCUCCCCC-UCUCUGGCUCGCCAGCUCUCCA--------GAUCCCGCAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAA---GUGCCUCGUUCUCG --........-....(((....)))((.((.((--------(......(((.....)))............(((((((((......))))))))).))).)---).)).......... ( -21.10, z-score = -1.77, R) >dp4.chr3 13054837 94 - 19779522 -----------UCCCUCUCGAGCCA--CAUCCA--------GAUCC---AGAACUCUGAAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGUGGUGCAGCGUUCUCC -----------....(((.((....--..)).)--------))...---(((((.(((..((((((.....(((((((((......)))))))))..))))))....))).))))).. ( -23.20, z-score = -2.06, R) >droPer1.super_2 473086 94 + 9036312 -----------UCCCUCUCGAGCCA--CAUCCA--------GAUCC---AGUACUCUGAAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGUGGUGCAGCGUUCUCC -----------.......((.((((--(...((--------(...(---((....))).............(((((((((......))))))))).)))..)))))....))...... ( -22.00, z-score = -1.71, R) >consensus __UCUCCCCC_UCCCUGGCUCGCCAGCUCUCCA________GAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAA___GUGCCUCGUUCUCG ................(((......................((((.....)))).................(((((((((......)))))))))...........)))......... (-12.31 = -13.31 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:04 2011