
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,880,803 – 15,880,862 |
| Length | 59 |
| Max. P | 0.545734 |

| Location | 15,880,803 – 15,880,862 |
|---|---|
| Length | 59 |
| Sequences | 11 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.62971 |
| G+C content | 0.40118 |
| Mean single sequence MFE | -10.74 |
| Consensus MFE | -5.56 |
| Energy contribution | -5.20 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.545734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15880803 59 - 21146708 ------ACAUCAUGGUGGUGCGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGUGUUUUC--- ------....((((..(((((((....)))))))..(((((((.....))))))).)))).....--- ( -13.30, z-score = -1.50, R) >droAna3.scaffold_13266 10430008 59 + 19884421 ------GCAUUCUCGUGGUGUGAAUCUUUGCACUUGUCUUUGAAUUAAUUAGUGAGCUUGUCUUU--- ------(((..((((.((((..(....)..)))).....((((.....))))))))..)))....--- ( -7.70, z-score = 0.49, R) >droEre2.scaffold_4845 10032191 59 - 22589142 ------UCAUCGUGGUGGUGUGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGUGUUUUC--- ------.((.(((...((((..(....)..))))..(((((((.....)))))))))))).....--- ( -10.60, z-score = -0.84, R) >droYak2.chr2R 7807782 52 + 21139217 -------------GAUGGUGUGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGCGUUUUC--- -------------...((((..(....)..))))..(((((((.....)))))))..........--- ( -8.60, z-score = -0.39, R) >droSec1.super_1 13358649 59 - 14215200 ------UCAUCGUGGUGGUGCGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGUGUUUUC--- ------.((.(((...(((((((....)))))))..(((((((.....)))))))))))).....--- ( -12.80, z-score = -1.31, R) >droSim1.chr2R 14528311 59 - 19596830 ------UCAUCGUGGUGGUGCGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGUGUUUUC--- ------.((.(((...(((((((....)))))))..(((((((.....)))))))))))).....--- ( -12.80, z-score = -1.31, R) >droPer1.super_2 423298 51 + 9036312 -----------------GUGUGAAUCUUUGCACUUGGCUUUGAAUUAAUUAGUGAGGAGACACUCUCC -----------------(.(((..((((..((((................))))..))))))).)... ( -8.79, z-score = 0.01, R) >dp4.chr3 13006474 51 - 19779522 -----------------GUGUGAAUCUUUGCACUUGGCUUUGAAUUAAUUAGUGAGGAGACACUCACC -----------------(((..(....)..)))..................(((((......))))). ( -10.90, z-score = -0.95, R) >droWil1.scaffold_180701 2184023 60 - 3904529 -----UGCUGUUGGUUGGUGUGAAUCUUUGCGCCUGGAUUUGAAUUAAUUAGUGAGUGCCUUCAU--- -----.(((((((((((((.(((((((........))))))).)))))))))).)))........--- ( -9.20, z-score = 0.60, R) >droMoj3.scaffold_6496 21394390 65 - 26866924 GCCCACUGCAUUUCAUGGUGUGAAUCUUUGCGCCCGUAUCUGCAUAAAUUAGUGCGAAUAUACUC--- ((.....)).......((((..(....)..)))).((((.(((((......))))).))))....--- ( -12.90, z-score = -0.57, R) >droVir3.scaffold_12875 16905752 54 + 20611582 --------GGCCAUUUGGUGUGAAUCUUUGCGCCCGUCUCUGAAUUAAUUA--GUGCCUGUUGU---- --------(((.....((((..(....)..)))).....((((.....)))--).)))......---- ( -10.50, z-score = -0.82, R) >consensus _______CAUC_UGGUGGUGUGAAUCUUUGCACUUGUCUUUGAAUUAAUUAAAGAGCGUGUUUUC___ ................(((((((....))))))).................................. ( -5.56 = -5.20 + -0.36)
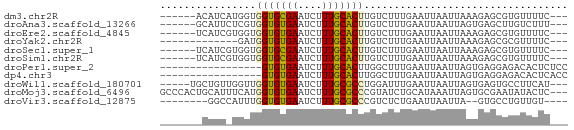
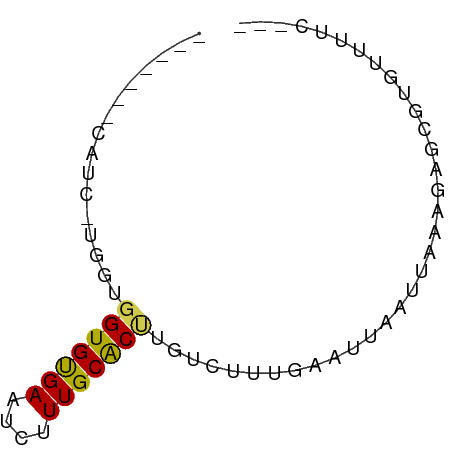
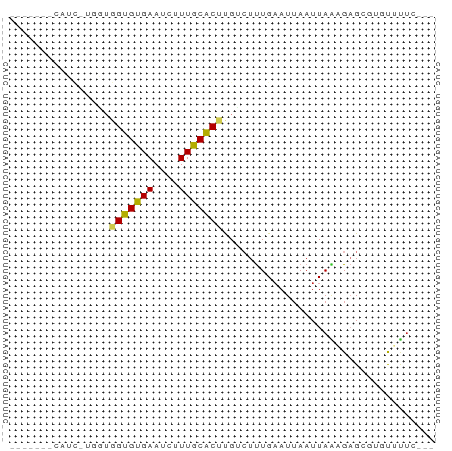
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:56 2011