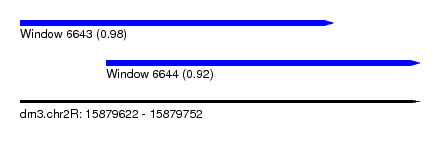
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,879,622 – 15,879,752 |
| Length | 130 |
| Max. P | 0.977757 |

| Location | 15,879,622 – 15,879,724 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Shannon entropy | 0.51615 |
| G+C content | 0.51958 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
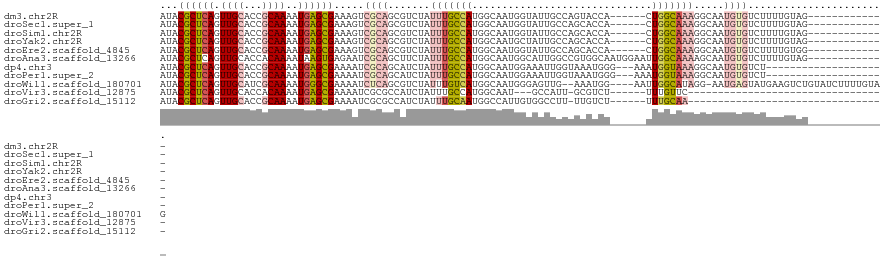
>dm3.chr2R 15879622 102 + 21146708 ---UUCCAGGCACU---UUGCCCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUA-UUGCCAGUACC------ ---.....((((..---.((((...((((((((....((((((.((((...))))..)))))).....))))))))....((((....)))).))))-.))))......------ ( -37.80, z-score = -2.92, R) >droSec1.super_1 13356541 102 + 14215200 ---UUCCAGGCACU---UUUCCCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUA-UUGCCAGCACC------ ---.....((((..---........((((((((....((((((.((((...))))..)))))).....)))))))).....((((((....))))))-.))))......------ ( -35.90, z-score = -2.71, R) >droSim1.chr2R 14526181 102 + 19596830 ---UUCCAGGCACU---UUGCCCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUA-UUGCCAGCACC------ ---.....((((..---.((((...((((((((....((((((.((((...))))..)))))).....))))))))....((((....)))).))))-.))))......------ ( -37.80, z-score = -2.62, R) >droYak2.chr2R 7806901 101 - 21139217 ---UUCCAGGCGCU---UUGCCCC-GCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGCUA-UUGCCAGCACC------ ---.....(((...---..)))..-((((((((....((((((.((((...))))..)))))).....)))))))).....(((..(((((((...)-)))))))))..------ ( -35.80, z-score = -1.70, R) >droEre2.scaffold_4845 10030048 102 + 22589142 ---UUCCAGGCGCU---UUGCCCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUA-UUGCCAGCACC------ ---.....((((..---.((((...((((((((....((((((.((((...))))..)))))).....))))))))....((((....)))).))))-.))))......------ ( -37.10, z-score = -1.91, R) >droAna3.scaffold_13266 10427399 108 - 19884421 ---UUCCAGGCACU---UUGCCCCCGCGUUGCGUAUACGCUCAGUUGCACCACAAAAUAAGUGAGAAUCGCAGCUUCUAUUUGCCAUGGCAAUGGCA-UUGGCCGUGGCAAUGGA ---.(((((((...---..))).....((((((......((((.(((..........))).))))...))))))......((((((((((.......-...)))))))))))))) ( -36.40, z-score = -0.96, R) >dp4.chr3 13005742 104 + 19779522 ------CAAGCACCGG-UCUACCCCGUGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCAUCUAUUUGCCAUGGCAAUGGAAAUUGGUAAAUGGGA---- ------......((.(-(.((((...(((((((....((((((.((((...))))..)))))).....)))))))((((((.((....))))))))....)))).)).)).---- ( -36.80, z-score = -2.52, R) >droPer1.super_2 422569 104 - 9036312 ------CAGGCACCGG-UCUACCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCAUCUAUUUGCCAUGGCAAUGGAAAUUGGUAAAUGGGA---- ------......((.(-(.((((....((((((....((((((.((((...))))..)))))).....)))))).((((((.((....))))))))....)))).)).)).---- ( -36.40, z-score = -1.85, R) >droWil1.scaffold_180701 2179732 106 + 3904529 -UUCAACUCAU-UCCAACCUCCCGCAUAUUGCGCAUACGCUCAGUUGCAUCGCAAAAUGGGCGAAAAUCUCAGCGUCUAUUUGUCAUGGCAAUGGGAGUUGAAAUGGA------- -......((((-(.((((.(((((((.((((.((....)).)))))))...((.(((((((((..........)))))))))......))...)))))))).))))).------- ( -31.90, z-score = -1.99, R) >droVir3.scaffold_12875 16900662 94 - 20611582 -UUCAAAAGGU-CUCACACUGUUGCACGUUGCGUAUACGCUCAGUUGCACCACAAAAUGAGCGAAAAUCGCGCCAUCUAUUUGCCAUGGCAAUGCC------------------- -.......(((-.(((...((((((((...(((....)))...).))))..)))...)))(((.....))))))......((((....))))....------------------- ( -21.10, z-score = 0.26, R) >droGri2.scaffold_15112 2467619 99 + 5172618 UCCCAUCACGUGCUCACACUGUUGCACGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCGCCAUCUAUUUGCAAUGGCCAUUGUGGC---------------- ..((((.((((((..........)))))).(((....((((((.((((...))))..)))))).....)))(((((.........)))))....)))).---------------- ( -30.00, z-score = -0.95, R) >consensus ___UUCCAGGCACU___UUUCCCCCGCGUUGCGUAUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCGUCUAUUUGCCAUGGCAAUGGUA_UUGCCAGCACC______ ...........................((((((....((((((.((((...))))..)))))).....))))))......((((....))))....................... (-20.58 = -20.92 + 0.33)


| Location | 15,879,650 – 15,879,752 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.51499 |
| G+C content | 0.46211 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
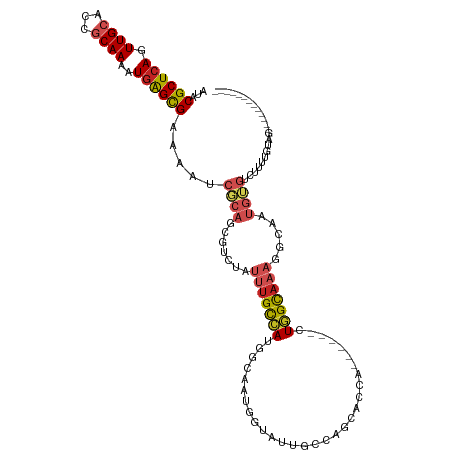
>dm3.chr2R 15879650 102 + 21146708 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUAUUGCCAGUACCA------CUGGCAAAGGCAAUGUGUCUUUUGUAG------------- .((((((((.((((...))))..)))))(((((.((((..((((...((((((....))))))(((((((.....------)))))))))))..)))).)))))))).------------- ( -40.90, z-score = -3.92, R) >droSec1.super_1 13356569 102 + 14215200 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUAUUGCCAGCACCA------CUGGCAAAGGCAAUGUGUCUUUUGUAG------------- .((((((((.((((...))))..)))))(((((.((((..((((...((((((....))))))(((((((.....------)))))))))))..)))).)))))))).------------- ( -40.10, z-score = -3.32, R) >droSim1.chr2R 14526209 102 + 19596830 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUAUUGCCAGCACCA------CUGGCAAAGGCAAUGUGUCUUUUGUAG------------- .((((((((.((((...))))..)))))(((((.((((..((((...((((((....))))))(((((((.....------)))))))))))..)))).)))))))).------------- ( -40.10, z-score = -3.32, R) >droYak2.chr2R 7806928 102 - 21139217 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGCUAUUGCCAGCACCA------CUGGCAAAGGCAAUGUGUCUUUUGUAG------------- .((((((((.((((...))))..)))))(((((.((((........((((....))))((((.(((((((.....------))))))).)))).)))).)))))))).------------- ( -39.00, z-score = -3.01, R) >droEre2.scaffold_4845 10030076 102 + 22589142 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAGUCGCAGCGUCUAUUUGCCAUGGCAAUGGUAUUGCCAGCACCA------CUGGCAAAGGCAAUGUGUCUUUUGUGG------------- .((((((((.((((...))))..)))))(((((.((((..((((...((((((....))))))(((((((.....------)))))))))))..)))).)))))))).------------- ( -39.80, z-score = -2.73, R) >droAna3.scaffold_13266 10427427 108 - 19884421 AUACGCUCAGUUGCACCACAAAAUAAGUGAGAAUCGCAGCUUCUAUUUGCCAUGGCAAUGGCAUUGGCCGUGGCAAUGGAAUUGGCAAAAGCAAUGUGUCUUUUGUAG------------- ..((((...(((((..(((.......)))......((...(((((((.((((((((..........)))))))))))))))...))....))))))))).........------------- ( -32.30, z-score = -0.87, R) >dp4.chr3 13005769 98 + 19779522 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCAUCUAUUUGCCAUGGCAAUGGAAAUUGGUAAAUGGG---AAAUGGUAAAGGCAAUGUGUCU-------------------- ...((((((.((((...))))..)))))).....((((((..((((((.((((..((((....))))....)))).---)))))).....))..))))...-------------------- ( -28.70, z-score = -1.72, R) >droPer1.super_2 422596 98 - 9036312 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCAUCUAUUUGCCAUGGCAAUGGAAAUUGGUAAAUGGG---AAAUGGUAAAGGCAAUGUGUCU-------------------- ...((((((.((((...))))..)))))).....((((((..((((((.((((..((((....))))....)))).---)))))).....))..))))...-------------------- ( -28.70, z-score = -1.72, R) >droWil1.scaffold_180701 2179764 114 + 3904529 AUACGCUCAGUUGCAUCGCAAAAUGGGCGAAAAUCUCAGCGUCUAUUUGUCAUGGCAAUGGGAGUUG--AAAUGG----AAUUGGCAUAGG-AAUGAGUAUGAAGUCUGUAUCUUUUGUAG .(((((.(((((.((((((.......)))).....(((((..(((((.((....)))))))..))))--)..)).----)))))))(((((-.............))))).......))). ( -25.62, z-score = 0.19, R) >droVir3.scaffold_12875 16900694 78 - 20611582 AUACGCUCAGUUGCACCACAAAAUGAGCGAAAAUCGCGCCAUCUAUUUGCCAUGGCAAU---GCCAUU-GCGUCU------UUUGUUC--------------------------------- ...((((((.(((.....)))..)))))).(((..((((.......((((....)))).---......-))))..------)))....--------------------------------- ( -19.04, z-score = -0.91, R) >droGri2.scaffold_15112 2467653 81 + 5172618 AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCGCCAUCUAUUUGCAAUGGCCAUUGUGGCCUU-UUGUCU------UUUGCAA--------------------------------- ...((((((.((((...))))..)))))).................((((((.((((.....))))..-......------.))))))--------------------------------- ( -22.00, z-score = -0.86, R) >consensus AUACGCUCAGUUGCACCGCAAAAUGAGCGAAAAUCGCAGCGUCUAUUUGCCAUGGCAAUGGUAUUGCCAGCACCA______CUGGCAAAGGCAAUGUGUCUUUUGUAG_____________ ...((((((.((((...))))..)))))).....((((.......(((((((..............................))))))).....))))....................... (-14.88 = -15.70 + 0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:55 2011