| Sequence ID | dm3.chr2R |
|---|---|
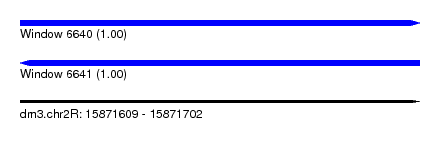
| Location | 15,871,609 – 15,871,702 |
| Length | 93 |
| Max. P | 0.997663 |

| Location | 15,871,609 – 15,871,702 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.50025 |
| G+C content | 0.39880 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
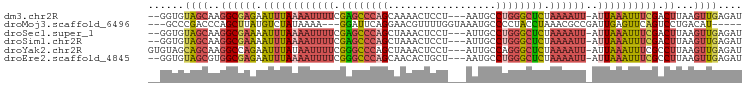
>dm3.chr2R 15871609 93 + 21146708 --GGUGUAGCAAGGCGAGAAUUUAAAAUUUUCGAGCCCAGCAAAACUCCU---AAUGCCUGGGCUCUAAAAUU-AUUAAAUUUCGACUUAAGUUGAGAU --....(((((((.(((((.(((((((((((.((((((((((........---..)).)))))))).))))))-.)))))))))).)))..)))).... ( -26.80, z-score = -3.04, R) >droMoj3.scaffold_6496 21378962 88 + 26866924 ---GCCCGACCCAGCUUAUGUCUAUAAAA---GGAUUCAGGAACGUUUUGGUAAAUGCCCCUACCUAAACGCCGAUUGAGUUCAGUCCUGACAU----- ---..............(((((.......---((((((((...(((((.((((........)))).)))))....))))))))......)))))----- ( -19.52, z-score = -1.42, R) >droSec1.super_1 13348588 93 + 14215200 --GGUGUAGCAAGGCGAAAAUUUAAAAUUUUCGAGCCCAGCUAAACUCCU---AUUGCCUGGGCUCUAAAAUU-AUUAAAUUUCGACUUAAGUUGAGAU --....(((((((.(((((.(((((((((((.((((((((.(((......---.))).)))))))).))))))-.)))))))))).)))..)))).... ( -26.40, z-score = -3.08, R) >droSim1.chr2R 14518239 93 + 19596830 --GGUGUAGCAAGGCGAAAAUUUAAAAUUUUCGAGCCCAGCUAAACUCCU---AUUGCCUGGGCUCUAAAAUU-AUUAAAUUUCGACUUAAGUUGAGAU --....(((((((.(((((.(((((((((((.((((((((.(((......---.))).)))))))).))))))-.)))))))))).)))..)))).... ( -26.40, z-score = -3.08, R) >droYak2.chr2R 7798427 95 - 21139217 GUGUAGCAGCAAGGCCAGAAUUUAUAAUUUUCGGGCCCAGCUAAACUCCU---AUUGCCAGGGCUCUAAAAUU-AUUAAAUUUCGCCUUAAGUUGAGAU ..((..(((((((((..((((((((((((((.((((((.((.........---...))..)))))).))))))-).))))))).)))))..))))..)) ( -28.80, z-score = -2.67, R) >droEre2.scaffold_4845 10022194 93 + 22589142 --GGUGUAGCGUGGCGAGAAUUUAAAAUUUUCGGGCCCAGCAACACUGCU---AAUGCCUGGGCUCUAAAAUU-AUUAAAUUUCGCCUUAAGUUGAGAU --....((((..(((((((.(((((((((((.((((((((((........---..)).)))))))).))))))-.))))))))))))....)))).... ( -32.10, z-score = -3.44, R) >consensus __GGUGUAGCAAGGCGAAAAUUUAAAAUUUUCGAGCCCAGCAAAACUCCU___AAUGCCUGGGCUCUAAAAUU_AUUAAAUUUCGACUUAAGUUGAGAU ......((((..(((((((.((...((((((.((((((((..................)))))))).))))))....)).))))).))...)))).... (-14.74 = -15.94 + 1.20)

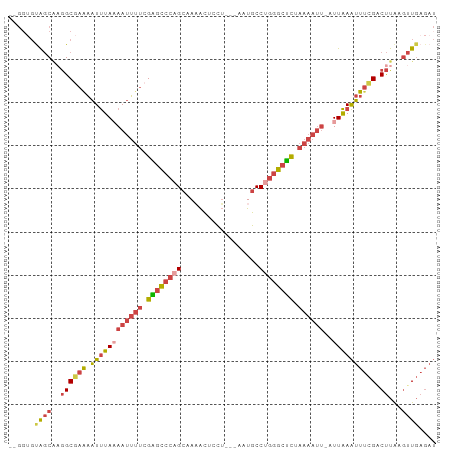
| Location | 15,871,609 – 15,871,702 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.50025 |
| G+C content | 0.39880 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -13.92 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
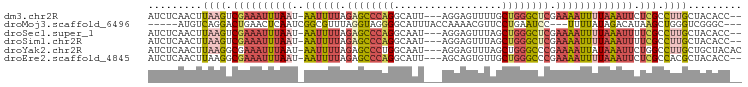
>dm3.chr2R 15871609 93 - 21146708 AUCUCAACUUAAGUCGAAAUUUAAU-AAUUUUAGAGCCCAGGCAUU---AGGAGUUUUGCUGGGCUCGAAAAUUUUAAAUUCUCGCCUUGCUACACC-- .........((((.((((((((((.-((((((.((((((((.((..---........)))))))))).))))))))))))).))).)))).......-- ( -27.40, z-score = -3.72, R) >droMoj3.scaffold_6496 21378962 88 - 26866924 -----AUGUCAGGACUGAACUCAAUCGGCGUUUAGGUAGGGGCAUUUACCAAAACGUUCCUGAAUCC---UUUUAUAGACAUAAGCUGGGUCGGGC--- -----...((((((((((......))))(((((.((((((....)))))).))))).))))))((((---.(((((....)))))..)))).....--- ( -22.00, z-score = -0.51, R) >droSec1.super_1 13348588 93 - 14215200 AUCUCAACUUAAGUCGAAAUUUAAU-AAUUUUAGAGCCCAGGCAAU---AGGAGUUUAGCUGGGCUCGAAAAUUUUAAAUUUUCGCCUUGCUACACC-- .........((((.((((((((((.-((((((.((((((((.....---..........)))))))).))))))))))).))))).)))).......-- ( -25.56, z-score = -3.17, R) >droSim1.chr2R 14518239 93 - 19596830 AUCUCAACUUAAGUCGAAAUUUAAU-AAUUUUAGAGCCCAGGCAAU---AGGAGUUUAGCUGGGCUCGAAAAUUUUAAAUUUUCGCCUUGCUACACC-- .........((((.((((((((((.-((((((.((((((((.....---..........)))))))).))))))))))).))))).)))).......-- ( -25.56, z-score = -3.17, R) >droYak2.chr2R 7798427 95 + 21139217 AUCUCAACUUAAGGCGAAAUUUAAU-AAUUUUAGAGCCCUGGCAAU---AGGAGUUUAGCUGGGCCCGAAAAUUAUAAAUUCUGGCCUUGCUGCUACAC .........((((((.(((((((.(-((((((.(.((((.(((...---.........))))))).).))))))))))))).).))))))......... ( -23.90, z-score = -1.00, R) >droEre2.scaffold_4845 10022194 93 - 22589142 AUCUCAACUUAAGGCGAAAUUUAAU-AAUUUUAGAGCCCAGGCAUU---AGCAGUGUUGCUGGGCCCGAAAAUUUUAAAUUCUCGCCACGCUACACC-- ............((((((((((((.-((((((.(.((((((((((.---....))))..)))))).).))))))))))))).)))))..........-- ( -28.80, z-score = -3.76, R) >consensus AUCUCAACUUAAGUCGAAAUUUAAU_AAUUUUAGAGCCCAGGCAAU___AGGAGUUUAGCUGGGCCCGAAAAUUUUAAAUUCUCGCCUUGCUACACC__ .........((((.(((((((((...((((((.((((((((..................)))))))).)))))).)))))).))).))))......... (-13.92 = -15.20 + 1.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:53 2011