| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,381,947 – 3,382,040 |
| Length | 93 |
| Max. P | 0.605415 |

| Location | 3,381,947 – 3,382,040 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.62 |
| Shannon entropy | 0.74019 |
| G+C content | 0.51204 |
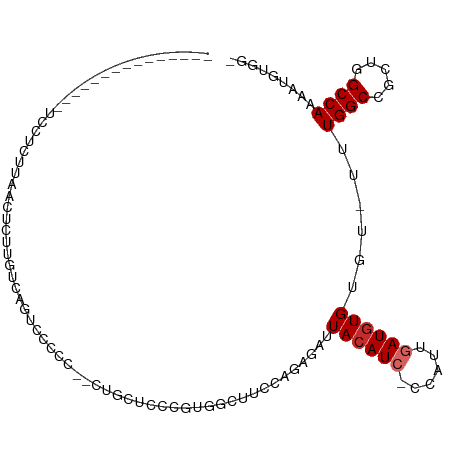
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -8.61 |
| Energy contribution | -8.94 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
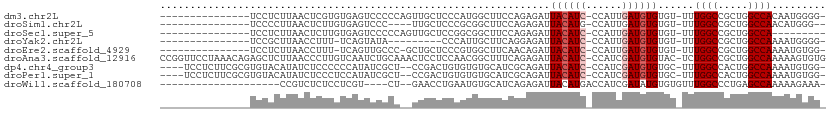
>dm3.chr2L 3381947 93 + 23011544 ---------------UCCUCUUAACUCGUGUGAGUCCCCCAGUUGCUCCCAUGGCUUCCAGAGAUUACAUC-CCAUUGAUGUGUGU-UUUGGCCGCUGGCCACAAUGGGG- ---------------........((((....)))).(((((.(((...(((((((...(((((..((((((-.....))))))..)-)))))))).)))...))))))))- ( -27.00, z-score = -0.71, R) >droSim1.chr2L 3320627 88 + 22036055 ---------------UCCCCUUAACUCUUGUGAGUCCC----UUGCUCCCGCGGCUUCCAGAGAUUACAUG-CCAUUGAUGUGUGU-UUUGGCCGCUGGCCAACAUGGG-- ---------------........((((....))))(((----.((..((.(((((...(((((..(((((.-......)))))..)-))))))))).))....)).)))-- ( -23.50, z-score = -0.38, R) >droSec1.super_5 1528967 85 + 5866729 ---------------UCCUCUUAACUCUUGUGAGUCCCCCAGUUGCUCCGGCGGCUUCCAGAGAUUACAUC-CCAUUGAUGUGUGU-UUUGGCCGCUGGCCA--------- ---------------........((((....))))............((((((((...(((((..((((((-.....))))))..)-))))))))))))...--------- ( -26.00, z-score = -1.89, R) >droYak2.chr2L 3366846 83 + 22324452 ---------------UCCGCUUAACCUUU-UCAGUAUA---------CCCAUUGCUUCAGGAGAUUACAUC-CCAUUGAUGUGUGU-UUUGGCCGCUGGCCAAAAUGGGG- ---------------..............-........---------(((...............((((((-.....)))))).((-(((((((...)))))))))))).- ( -20.70, z-score = -0.34, R) >droEre2.scaffold_4929 3411833 91 + 26641161 ---------------UCCUCUUAACCUUU-UCAGUUGCCC-GCUGCUCCCGUGGCUUCAACAGAUUACAUC-CCAUUGAUGUGUGU-UUUGGCCGCUGGCCAAAAUGUGG- ---------------..............-...((((...-((..(....)..))..))))....((((((-.....)))))).((-(((((((...)))))))))....- ( -23.60, z-score = -1.40, R) >droAna3.scaffold_12916 2837819 109 - 16180835 CCGGUUCCUAAACAGAGCUCUUAACCCUUGUCAAUCUGCAAACUCCUCCAACGGCUUUCAGAGAUUACAUC-CCAUCGAUGUGUAC-UCUGGCCGCUGGCCAAAAAGUGUG ..(((((.......)))))..................(((.......(((.((((...(((((..((((((-.....))))))..)-)))))))).)))........))). ( -25.56, z-score = -1.41, R) >dp4.chr4_group3 5511653 102 - 11692001 ----UCCUCUUCGCGUGUACAUAUCUCCCCCCAUAUCGCU--CCGACUGUGUGUGCAUCGCAGAUUACAUC-CCAUCGAUGUGUGC-UUUGGCCACUGGCCAAAAUGUGG- ----.......((((((((((((((..........(((..--.)))(((((.......)))))........-.....)))))))))-(((((((...)))))))))))).- ( -29.10, z-score = -2.20, R) >droPer1.super_1 2608735 102 - 10282868 ----UCCUCUUCGCGUGUACAUAUCUCCCUCCAUAUCGCU--CCGACUGUGUGUGCAUCGCAGAUUACAUC-CCAUCGAUGUGUGC-UUUGGCCACUGGCCAAAAUGUGG- ----.......((((((((((((((..........(((..--.)))(((((.......)))))........-.....)))))))))-(((((((...)))))))))))).- ( -29.10, z-score = -2.18, R) >droWil1.scaffold_180708 1680740 84 - 12563649 --------------------CCGUCUCUCCUCGU----CU--GAACCUGAAUGUGCAUCAGAGAUUACAUGACCAUCGAUAUGUGUGUUUGGCCCUGAGCCAAAAAGAAA- --------------------..(((.....((((----.(--((.(((((.......)))).).))).)))).....))).......((((((.....))))))......- ( -12.80, z-score = 0.84, R) >consensus _______________UCCUCUUAACUCUUGUCAGUCCCCC__CUGCUCCCGUGGCUUCCAGAGAUUACAUC_CCAUUGAUGUGUGU_UUUGGCCGCUGGCCAAAAUGUGG_ .................................................................((((((......))))))......((((.....))))......... ( -8.61 = -8.94 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:42 2011