
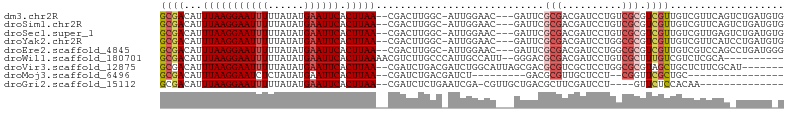
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,790,472 – 15,790,570 |
| Length | 98 |
| Max. P | 0.530549 |

| Location | 15,790,472 – 15,790,570 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.89 |
| Shannon entropy | 0.55136 |
| G+C content | 0.44761 |
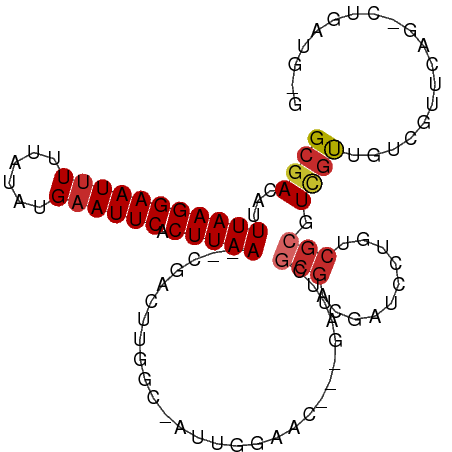
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -8.81 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15790472 98 + 21146708 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGACUUGGC-AUUGGAAC---GAUUCGCGACGAUCCUGUCGCGUCGUUGUCGUUCAGUCUGAUGUG ..(((..(((((((((((......)))))).)))))--.........-....((((---(((.(((((((....))))))).....))))))).)))....... ( -29.50, z-score = -1.83, R) >droSim1.chr2R 14449695 98 + 19596830 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGACUUGGC-AUUGGAAC---GAUUCGCGACGAUCCUGUCGCGUCGUUGUCGUUCAGUCUGAUGUG ..(((..(((((((((((......)))))).)))))--.........-....((((---(((.(((((((....))))))).....))))))).)))....... ( -29.50, z-score = -1.83, R) >droSec1.super_1 13280087 98 + 14215200 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGACUUGGC-AUUGGAAC---GAUUCGCGACGAUCCUGUCGCGUCGUUGUCGUUGAGUCUGAUGUG ...(((((((((((((((......)))))).)))).--.((((..((-...(.(((---((..(((((((....)))))))))))).).))..)))).))))). ( -33.10, z-score = -2.96, R) >droYak2.chr2R 7726068 98 - 21139217 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGACUUGGC-AUUGGAAC---GAUUCGCGACGAUCCUGGCGCGUCGUUGUCGUUCAUCCUGAUGUG .......(((((((((((......)))))).)))))--.......((-(((.((((---(((..((((((.........)))))).))))))).....))))). ( -26.20, z-score = -0.93, R) >droEre2.scaffold_4845 9952763 98 + 22589142 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGACUUGGC-AUUGGAAC---GAUUCGCGACGAUCCUGGCGCGUCGUUGUCGUCCAGCCUGAUGGG ((((((..((((((((((......)))))).))))(--((((.((.(-(..(((.(---(........)).))))).)).)))))))))))(((......))). ( -28.50, z-score = -1.30, R) >droWil1.scaffold_180701 1850006 92 - 3904529 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAAAACGUCUUGCCCAUUGCCAUU--GGGACGCGACGAUCCUGUCGCUUUGUCGUCUCGCA---------- ((((((((((((((((((......)))))).)))))).((((.((((((.......)--))).)).))))....))))))..............---------- ( -29.90, z-score = -3.76, R) >droVir3.scaffold_12875 16797227 95 - 20611582 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGAUCUGACGAUCUGGCAUUAGCGACGCGUCGCUCCUGGCGCGUAGCUGCUCUUCGCAU------- ((((...(((((((((((......)))))).)))))--.((((....)))).((((...((.((((((((....)))))))).))))))..))))..------- ( -31.60, z-score = -2.74, R) >droMoj3.scaffold_6496 21268852 75 + 26866924 GCGACAUUUAAGGAAUCUCUAUAUGAAUUCACUUAA--CGAUCUGACGAUCU---------GACGCGUUGCUCCU--CGGUUCGCUGC---------------- (((((..(((((((((.((.....)))))).)))))--.((((....)))).---------.....)))))....--(((....))).---------------- ( -15.90, z-score = -0.57, R) >droGri2.scaffold_15112 2371887 83 + 5172618 GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA--CGAUCUCUGAAUCGA-CGUUGCUGACGCUUCGAUCCU----GUUCUCCACAA-------------- (((((..(((((((((((......)))))).)))))--((((......)))).-.)))))...............----...........-------------- ( -16.10, z-score = -1.41, R) >consensus GCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAA__CGACUUGGC_AUUGGAAC___GAUUCGCGACGAUCCUGUCGCGUCGUUGUCGUUCAG_CUGAUG_G ((((...(((((((((((......)))))).)))))............................(((.((....)).))).))))................... ( -8.81 = -9.50 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:43 2011