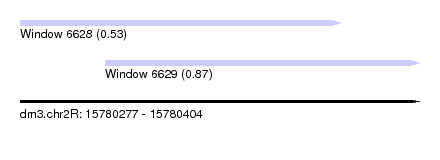
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,780,277 – 15,780,404 |
| Length | 127 |
| Max. P | 0.868314 |

| Location | 15,780,277 – 15,780,379 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.19 |
| Shannon entropy | 0.56541 |
| G+C content | 0.46784 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
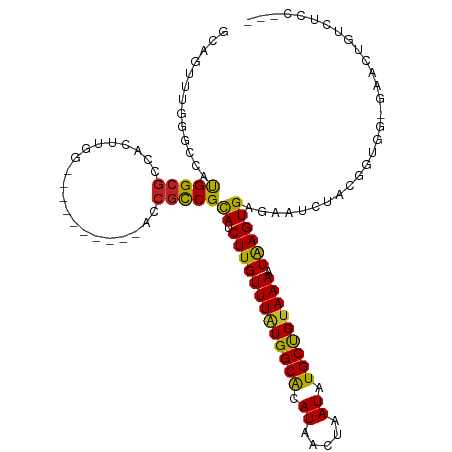
>dm3.chr2R 15780277 102 + 21146708 GCAGUUUUGGUCCUGGCGCCACUUGG----------ACCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAACUGUCUCC--- (((((((.(((((.((.....)).))----------)))(((((.(((..(((((.(((((((.....)))).)))...)))))..))).......))))))))))))....--- ( -34.30, z-score = -2.85, R) >droVir3.scaffold_12875 16780417 85 - 20611582 ---GGUAGCUCCGUGGCGCCGCUUUG----------GCCAUCGCAUCUUGUUUGUGGCGAAUAACUAAUAUGCUGUAAAAUGAGUGUGAUACUCGAGC----------------- ---(((.(((....))))))((((.(----------(..((((((.((..((((..(((.((.....)).)))..).)))..)))))))).)).))))----------------- ( -23.70, z-score = -0.40, R) >droMoj3.scaffold_6496 21242404 84 + 26866924 ----GCAGCUACGUGGCGCCGCAUUG----------GCCGCCGCAUCUUGUUUAUGGCGAAUAACUAAUAUGCCGUAAAAUAAGUGAGAAACUCUUGC----------------- ----((((....((((((((.....)----------).))))))..(((((((((((((.((.....)).)))))).)))))))..........))))----------------- ( -25.60, z-score = -1.26, R) >droGri2.scaffold_15112 2356219 87 + 5172618 ----GCAGCUCUGUGGCGCCGCAUUGGG-------CGCCGUCGCAUCUUGUUUAUGGCGAAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUCUAGAGU----------------- ----...((((((.((((((......))-------)))).((.((.(((((((((((((.((.....)).)))))).))))))))).))...))))))----------------- ( -29.70, z-score = -2.09, R) >dp4.chr3 12908748 101 + 19779522 GCA--UUGGGCCAUGGCGCCACUCCG----------UCCGUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACACAGGGAUUUCGCUUCCU-- ...--..((((.((((((......))----------.)))).)).....((((((((((.((.....)).)))))))))).((((((((.(((......))))))))))))).-- ( -25.80, z-score = -0.71, R) >droPer1.super_2 321971 101 - 9036312 GCA--UUGGGCCAUGGCGCCACUCCG----------UCCGUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACACAGGGAUUUUGCUUCCU-- ...--..((((.((((((......))----------.)))).)).....((((((((((.((.....)).)))))))))).((((.(((((((......))))))))))))).-- ( -23.90, z-score = -0.04, R) >droWil1.scaffold_180701 2036043 111 + 3904529 -CUUUUUUCCCUGUGGCGCCACAUUGCAUGGCGACCGUCGUCGCAUCUUGUUUAUGGCAAAUAACUAAUAUGCUGGAAAAUAAGUGAGAAUCCAAAAAAAAGUUUUUUUUCG--- -(((((((((.((((....))))..((((((((((....)))))...((((.....))))........))))).)))))).))).(((((................))))).--- ( -23.29, z-score = -0.37, R) >droSim1.chr2R 14439449 102 + 19596830 GCAGUUUUGGUCAUGGCGCCACUUGG----------ACCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAACUGUCUCC--- (((((((.((((.((....))....)----------)))(((((.(((..(((((.(((((((.....)))).)))...)))))..))).......))))))))))))....--- ( -31.00, z-score = -1.83, R) >droSec1.super_1 13269954 105 + 14215200 GCAGUUUUGGUCCUGGCGCCACUUGG----------ACCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAACAGUCGUCUCC ((.((...(((((.((.....)).))----------))))).)).(((..(((((.(((((((.....)))).)))...)))))..)))......((.(((((....))))).)) ( -30.40, z-score = -1.13, R) >droYak2.chr2R 7715624 102 - 21139217 GCAGUUUUGGUCCUGGCGCCACUUAG----------ACCGCCGUAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAACUGUCUCC--- (((((((((((......))))....(----------.(((((((((((..(((((.(((((((.....)))).)))...)))))..)))...))))))))))))))))....--- ( -33.70, z-score = -3.11, R) >droEre2.scaffold_4845 9942544 101 + 22589142 GCAGUUUGGCUCCUGGCGCCACUUAG----------ACCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGG-GGGGAACUGUCUCC--- ((((((((((.......)))......----------.((.(((..(((..(((((.(((((((.....)))).)))...)))))..))).....)))-.)))))))))....--- ( -29.90, z-score = -1.37, R) >droAna3.scaffold_13266 10341058 98 - 19884421 GCAUUUU--GCGACGGCGCCACUUGG----------AC-GUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACGGCCA-GACAUGCCCCC--- .((((((--((((((...((....))----------.)-))))))....((((((((((.((.....)).)))))))))).))))).........(((..-.....)))...--- ( -27.90, z-score = -1.70, R) >consensus GCAGUUUGGGCCAUGGCGCCACUUGG__________ACCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUCUACGGUGG_GAACUGUCUCC___ .............(((((....................)))))((.(((((((((((((.((.....)).))))))))).))))))............................. (-11.64 = -11.07 + -0.56)


| Location | 15,780,304 – 15,780,404 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.24 |
| Shannon entropy | 0.65021 |
| G+C content | 0.40406 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -9.27 |
| Energy contribution | -8.89 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
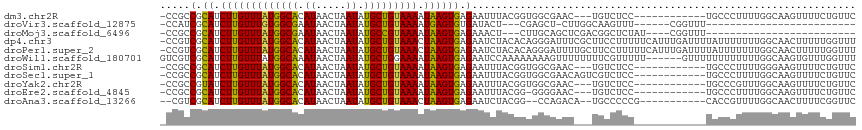
>dm3.chr2R 15780304 100 + 21146708 -CCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAAC---UGUCUCC------------UGCCCUUUUGGCAAGUUUUCUGUUC -((((((..(((..(((((.(((((((.....)))).)))...)))))..))).....)))))).((((---.(...(.------------((((.....)))).)....).)))) ( -27.10, z-score = -2.22, R) >droVir3.scaffold_12875 16780441 80 - 20611582 -CCAUCGCAUCUUGUUUGUGGCGAAUAACUAAUAUGCUGUAAAAUGAGUGUGAUACU---CGAGCU-CUUGGCAAGUUU------CGGUUU------------------------- -..((((((.((..((((..(((.((.....)).)))..).)))..))))))))(((---.(((((-.......)))))------.)))..------------------------- ( -17.40, z-score = -0.43, R) >droMoj3.scaffold_6496 21242427 83 + 26866924 -CCGCCGCAUCUUGUUUAUGGCGAAUAACUAAUAUGCCGUAAAAUAAGUGAGAAACU---CUUGCAGCUCGACGGCUCUAU----CGGUUU------------------------- -..(((((......(((((((((.((.....)).)))))))))....(..((.....---))..).....).)))).....----......------------------------- ( -20.80, z-score = -1.18, R) >dp4.chr3 12908773 115 + 19779522 -CCGUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACACAGGGAUUUCGCUUCCUUUUUCAUUUGAUUUUAUUUUUUUGGCAACUUUUUGGUUU -..(((.......((((((((((.((.....)).))))))))))((((((((((.......(((((......))))))))))))))).............)))((((....)))). ( -22.01, z-score = -1.22, R) >droPer1.super_2 321996 115 - 9036312 -CCGUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACACAGGGAUUUUGCUUCCUUUUUCAUUUGAUUUUAUUUUUUUGGCAACUUUUUGGUUU -..(((.......((((((((((.((.....)).))))))))))((((((((((.......(((((......))))))))))))))).............)))((((....)))). ( -22.01, z-score = -1.16, R) >droWil1.scaffold_180701 2036078 110 + 3904529 GUCGUCGCAUCUUGUUUAUGGCAAAUAACUAAUAUGCUGGAAAAUAAGUGAGAAUCCAAAAAAAAGUUUUUUUUCGUUUUU------GUUUUUUUUUUUUGGCAAGUGUUUGGUUU (((((.((.....))..)))))....((((((..(((..(((((.(((..(((((..((((((....))))))..))))).------.)))...)))))..))).....)))))). ( -20.90, z-score = -0.95, R) >droSim1.chr2R 14439476 100 + 19596830 -CCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAAC---UGUCUCC------------UGCCCUUUUGGGAAGUUUUCUGUUC -((((((..(((..(((((.(((((((.....)))).)))...)))))..))).....)))))).((((---.(....(------------(.(((....))).))....).)))) ( -27.90, z-score = -2.32, R) >droSec1.super_1 13269981 103 + 14215200 -CCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAACAGUCGUCUCC------------UGCCCUUUUGGCAAGUUUUCUGUUC -((((((..(((..(((((.(((((((.....)))).)))...)))))..))).....)))))).((((((....((..------------((((.....))))))....)))))) ( -31.50, z-score = -3.18, R) >droYak2.chr2R 7715651 100 - 21139217 -CCGCCGUAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGGUGGCGAAC---UGUCUCC------------UGCCCGUUUGGCAAGUUUUCUGUUC -(((((((((((..(((((.(((((((.....)))).)))...)))))..)))...)))))))).((((---.(...(.------------((((.....)))).)....).)))) ( -30.80, z-score = -3.48, R) >droEre2.scaffold_4845 9942571 99 + 22589142 -CCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUUUACGG-GGGGAAC---UGUCUCC------------UGCCCUUUUGGCAAGUUUUCUGUUC -..(((...(((..(((((.(((((((.....)))).)))...)))))..)))......((-(((((..---....)))------------).)))....)))............. ( -26.30, z-score = -1.35, R) >droAna3.scaffold_13266 10341083 99 - 19884421 --CGUCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAACUAAGUGAGAAUCUACGG--CCAGACA--UGCCCCCG-----------CACCGUUUUGGCAACUUUUCGGUUC --..((((.....((((((((((.((.....)).))))))))))...))))(((......(--((((((.--(((....)-----------))..).)))))).....)))..... ( -23.10, z-score = -0.64, R) >consensus _CCGCCGCAUCUUGUUUAUGGCACAUAACUAAUAUGCUGUAAAAUAAGUGAGAAUCUACGGUGGCGAAC___UGUCUCC____________UGCCCUUUUGGCAAGUUUUCUGUUC .....(.((.(((((((((((((.((.....)).))))))))).)))))).)................................................................ ( -9.27 = -8.89 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:42 2011