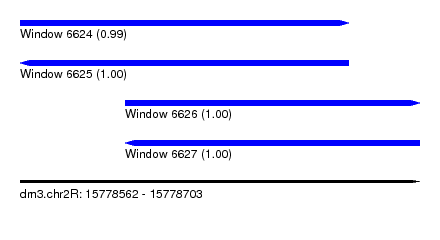
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,778,562 – 15,778,703 |
| Length | 141 |
| Max. P | 0.999771 |

| Location | 15,778,562 – 15,778,678 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.50414 |
| G+C content | 0.31895 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
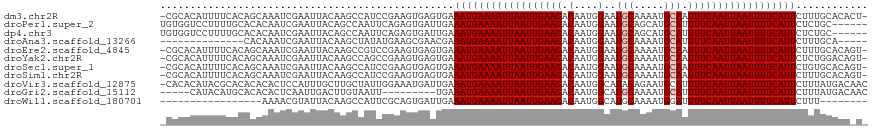
>dm3.chr2R 15778562 116 + 21146708 -CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCAUCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGCACACU- -.(((...((((((..((..(((..............)))..)).))))))((((((((((((((((.(....)..(((.....))).)))))))))))))))).....))).....- ( -25.34, z-score = -2.11, R) >droPer1.super_2 320024 112 - 9036312 UGUGGUCCUUUUGCACACAAUCGAAUUACAGCCAAUUCAGAGUGAUUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAGCAUGCAUUUUCAAUUAAUUUUCAUUCUCUGC------ ((((((.(..(((....)))..).)))))).......(((((.......((((((((((((((((........((((((.....))))))))))))))))))))))))))).------ ( -27.21, z-score = -1.60, R) >dp4.chr3 12906820 112 + 19779522 UGUGGUCCUUUUGCACACAAUCGAAUUACAGCCAAUUCAGAGUGAUUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAGCAUGCAUUUUCAAUUAAUUUUCAUUCUCUGC------ ((((((.(..(((....)))..).)))))).......(((((.......((((((((((((((((........((((((.....))))))))))))))))))))))))))).------ ( -27.21, z-score = -1.60, R) >droAna3.scaffold_13266 10339356 99 - 19884421 --------------CACAAAUCGAAUUACAAGCUAUAUGAAGCGAACGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAUUUUCAAUUAAUUUUCAUUCUUUGCA----- --------------...........................(((((.(((.((((((((((((((........((((((.....)))))))))))))))))))))))))))).----- ( -25.00, z-score = -3.87, R) >droEre2.scaffold_4845 9940876 116 + 22589142 -CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCGUCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGCACAGU- -.(((...((((((..((..(((.((........)).)))..)).))))))((((((((((((((((.(....)..(((.....))).)))))))))))))))).....))).....- ( -25.50, z-score = -1.60, R) >droYak2.chr2R 7713929 116 - 21139217 -CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCAGCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUCUGGACAGU- -.(..((.((((((..((..(((..............)))..)).))))))((((((((((((((((.(....)..(((.....))).)))))))))))))))).....))..)...- ( -22.74, z-score = -0.70, R) >droSec1.super_1 13268268 116 + 14215200 -CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCAUCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUGUGCACAGU- -.(((((.((((((..((..(((..............)))..)).))))))((((((((((((((((.(....)..(((.....))).))))))))))))))))...))))).....- ( -29.64, z-score = -2.93, R) >droSim1.chr2R 14437762 116 + 19596830 -CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCAUCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGCACAGU- -.(((...((((((..((..(((..............)))..)).))))))((((((((((((((((.(....)..(((.....))).)))))))))))))))).....))).....- ( -25.34, z-score = -1.81, R) >droVir3.scaffold_12875 16778187 117 - 20611582 -CACACAUACGCACACACACUCCAUUUGCUUGCUAUUGGAAAUGAUUGAAAUGAAAAUUAAUUGAAGACAAUGGCAUACAGAAUGCAUUUUCAAUUAAUUUUCAUUCUUUAUGACAAC -....((((.............(((((.(........).))))).....(((((((((((((((((((.....((((.....)))).)))))))))))))))))))...))))..... ( -26.00, z-score = -3.88, R) >droGri2.scaffold_15112 2353895 104 + 5172618 -----CAUACAUGCACACACUCAAUUGACUUGUAAUU---------UGAAAUGAAAAUUAAUUGAAGACAAUGGCAUGCAAAAUGCAUUUUCAAUUAAUUUUCAUUCUUUAUGACAAC -----........................((((....---------...(((((((((((((((((((.....((((.....)))).))))))))))))))))))).......)))). ( -23.64, z-score = -3.27, R) >droWil1.scaffold_180701 2034417 93 + 3904529 -----------------AAAACGUAUUACAAGCCAUUCGCAGUGAUUGAAAUGAAAAUUAAUUGAAGACAAUGGCAUGCAAAAUGGAUUUUCAAUUAAUUUUCAUUCUUU-------- -----------------....((.(((((..((.....)).))))))).(((((((((((((((((((..((.(....)...))...)))))))))))))))))))....-------- ( -19.10, z-score = -1.72, R) >consensus _CGCACAUUUUCACAGCAAAUCGAAUUACAAGCCAUCCGAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAUUUUCAAUUAAUUUUCAUUCUUUGCACA_U_ .................................................((((((((((((((((((.(....)..(((.....))).))))))))))))))))))............ (-15.46 = -15.65 + 0.18)
| Location | 15,778,562 – 15,778,678 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.50414 |
| G+C content | 0.31895 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.59 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15778562 116 - 21146708 -AGUGUGCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGGAUGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG- -...(..((.....(((((((((((((((..(((.....)))..........)))))))))))))))((((((.((..((((((........))))))..))..)))))).))..).- ( -32.00, z-score = -3.19, R) >droPer1.super_2 320024 112 + 9036312 ------GCAGAGAAUGAAAAUUAAUUGAAAAUGCAUGCUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCAAUCACUCUGAAUUGGCUGUAAUUCGAUUGUGUGCAAAAGGACCACA ------(((((((.((((((((((((((((((((.....)))))........)))))))))))))))))))...(((..(((((((....)))))))...))))))............ ( -28.50, z-score = -2.05, R) >dp4.chr3 12906820 112 - 19779522 ------GCAGAGAAUGAAAAUUAAUUGAAAAUGCAUGCUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCAAUCACUCUGAAUUGGCUGUAAUUCGAUUGUGUGCAAAAGGACCACA ------(((((((.((((((((((((((((((((.....)))))........)))))))))))))))))))...(((..(((((((....)))))))...))))))............ ( -28.50, z-score = -2.05, R) >droAna3.scaffold_13266 10339356 99 + 19884421 -----UGCAAAGAAUGAAAAUUAAUUGAAAAUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCGUUCGCUUCAUAUAGCUUGUAAUUCGAUUUGUG-------------- -----.((((((((((((((((((((((((((((.....)))))........)))))))))))))))))).....(((......)))...........))))).-------------- ( -25.20, z-score = -3.59, R) >droEre2.scaffold_4845 9940876 116 - 22589142 -ACUGUGCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGGACGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG- -...(..((.....(((((((((((((((..(((.....)))..........)))))))))))))))((((((.((..(((((..(....)..)))))..))..)))))).))..).- ( -29.50, z-score = -2.76, R) >droYak2.chr2R 7713929 116 + 21139217 -ACUGUCCAGAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGGCUGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG- -...(..((.....(((((((((((((((..(((.....)))..........)))))))))))))))((((((.((..(((..(........)..)))..))..)))))).))..).- ( -25.00, z-score = -1.30, R) >droSec1.super_1 13268268 116 - 14215200 -ACUGUGCACAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGGAUGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG- -.....(((((...(((((((((((((((..(((.....)))..........)))))))))))))))((((((.((..((((((........))))))..))..)))))).))))).- ( -34.80, z-score = -4.37, R) >droSim1.chr2R 14437762 116 - 19596830 -ACUGUGCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGGAUGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG- -...(..((.....(((((((((((((((..(((.....)))..........)))))))))))))))((((((.((..((((((........))))))..))..)))))).))..).- ( -32.00, z-score = -3.58, R) >droVir3.scaffold_12875 16778187 117 + 20611582 GUUGUCAUAAAGAAUGAAAAUUAAUUGAAAAUGCAUUCUGUAUGCCAUUGUCUUCAAUUAAUUUUCAUUUCAAUCAUUUCCAAUAGCAAGCAAAUGGAGUGUGUGUGCGUAUGUGUG- ((...((((..(((((((((((((((((((((((((.....))).))))...)))))))))))))))))).......(((((...(....)...)))))))))...)).........- ( -26.00, z-score = -1.73, R) >droGri2.scaffold_15112 2353895 104 - 5172618 GUUGUCAUAAAGAAUGAAAAUUAAUUGAAAAUGCAUUUUGCAUGCCAUUGUCUUCAAUUAAUUUUCAUUUCA---------AAUUACAAGUCAAUUGAGUGUGUGCAUGUAUG----- ..(((((((..(((((((((((((((((((((((((.....))).))))...))))))))))))))))))..---------.....(((.....)))..)))).)))......----- ( -22.30, z-score = -1.38, R) >droWil1.scaffold_180701 2034417 93 - 3904529 --------AAAGAAUGAAAAUUAAUUGAAAAUCCAUUUUGCAUGCCAUUGUCUUCAAUUAAUUUUCAUUUCAAUCACUGCGAAUGGCUUGUAAUACGUUUU----------------- --------...((((((((((((((((((..........(((......))).)))))))))))))))))).......(((((.....))))).........----------------- ( -16.80, z-score = -1.25, R) >consensus _A_UGUGCAAAGAAUGAAAAUUAAUUGAAAAUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUCGAAUGGCUUGUAAUUCGAUUUGCUGUGAAAAUGUGCG_ ...........((((((((((((((((((...(((...((.....)).))).))))))))))))))))))................................................ (-16.50 = -16.59 + 0.09)

| Location | 15,778,599 – 15,778,703 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Shannon entropy | 0.46645 |
| G+C content | 0.33657 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.26 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15778599 104 + 21146708 GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGC-ACACUGCGACCGCCGCACGUAC-AUACAUAC-------- ..((((.(..(((((((((((((((((((.(....)..(((.....))).)))))))))))))))))..))..)-.))))(((.....)))......-........-------- ( -27.10, z-score = -3.55, R) >droAna3.scaffold_13266 10339380 87 - 19884421 GAAGCGAACGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAUUUUCAAUUAAUUUUCAUUCUUUG---CAGAAGAAACGUGG------------------------ ...(((((.(((.((((((((((((((........((((((.....)))))))))))))))))))))))))))---).............------------------------ ( -25.00, z-score = -4.53, R) >droEre2.scaffold_4845 9940913 100 + 22589142 GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGC-ACAGUGCAACCACCGUACGUAC-AUAC------------ ...(((.....((((((((((((((((((.(....)..(((.....))).)))))))))))))))))).....)-)).((((.......))))....-....------------ ( -24.40, z-score = -2.93, R) >droYak2.chr2R 7713966 109 - 21139217 GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUCUGG-ACAGUGCGACCACCGCACGUACUACAUAUACUCGU---- .....(((((.((((((((((((((((((.(....)..(((.....))).))))))))))))))))))...(((-((.(((((.....))))))).)))....)))))..---- ( -31.00, z-score = -4.15, R) >droSec1.super_1 13268305 104 + 14215200 GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUGUGC-ACAGUGCGACUGCCGCACGUAC-AUACAUAC-------- ...(((..(..((((((((((((((((((.(....)..(((.....))).))))))))))))))))))..)..)-)).(((((.....)))))....-........-------- ( -30.30, z-score = -3.71, R) >droSim1.chr2R 14437799 104 + 19596830 GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGC-ACAGUGCGACCGCCGCACGUAC-AUACAUAC-------- ...(((.....((((((((((((((((((.(....)..(((.....))).)))))))))))))))))).....)-)).(((((.....)))))....-........-------- ( -29.10, z-score = -3.83, R) >droVir3.scaffold_12875 16778224 114 - 20611582 GAAAUGAUUGAAAUGAAAAUUAAUUGAAGACAAUGGCAUACAGAAUGCAUUUUCAAUUAAUUUUCAUUCUUUAUGACAACAUAUUCAUAUCCCCUCAGCCACAAGCACACACAC ....((..((((((((((((((((((((((.....((((.....)))).)))))))))))))))))))...(((((........))))).....)))..))............. ( -24.60, z-score = -4.13, R) >droGri2.scaffold_15112 2353926 106 + 5172618 -------UUGAAAUGAAAAUUAAUUGAAGACAAUGGCAUGCAAAAUGCAUUUUCAAUUAAUUUUCAUUCUUUAUGACAACAUAUUCAUAUCCUCUCCGC-ACACACACACACAC -------....(((((((((((((((((((.....((((.....)))).)))))))))))))))))))...(((((........)))))..........-.............. ( -23.20, z-score = -4.71, R) >consensus GAAGUGAGUGAAAUGAAAAUUAAUUGAAGACAAUGGAAUGCAAAAUGCAAUUUCAAUUAAUUUUCAUUCUUUGC_ACAGUGCAACCACCGCACGUAC_AUACAUAC________ .......(((.((((((((((((((((((.(....)..(((.....))).))))))))))))))))))...(((......)))..))).......................... (-15.95 = -15.26 + -0.69)

| Location | 15,778,599 – 15,778,703 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Shannon entropy | 0.46645 |
| G+C content | 0.33657 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -16.95 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15778599 104 - 21146708 --------GUAUGUAU-GUACGUGCGGCGGUCGCAGUGU-GCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC --------((..((.(-(((((((((.....)))).)))-)))..((((((((((((((((((..(((.....)))..........)))))))))))))))))).))..))... ( -28.50, z-score = -3.13, R) >droAna3.scaffold_13266 10339380 87 + 19884421 ------------------------CCACGUUUCUUCUG---CAAAGAAUGAAAAUUAAUUGAAAAUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCGUUCGCUUC ------------------------.............(---(((.(((((((((((((((((((((((.....)))))........))))))))))))))))))..)).))... ( -20.80, z-score = -4.24, R) >droEre2.scaffold_4845 9940913 100 - 22589142 ------------GUAU-GUACGUACGGUGGUUGCACUGU-GCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC ------------((..-((..((((((((....))))))-))...((((((((((((((((((..(((.....)))..........)))))))))))))))))).))..))... ( -28.90, z-score = -4.37, R) >droYak2.chr2R 7713966 109 + 21139217 ----ACGAGUAUAUGUAGUACGUGCGGUGGUCGCACUGU-CCAGAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC ----..((((...........(..(((((....))))).-.).((((.(((((((((((((((..(((.....)))..........)))))))))))))))))))))))..... ( -29.70, z-score = -3.32, R) >droSec1.super_1 13268305 104 - 14215200 --------GUAUGUAU-GUACGUGCGGCAGUCGCACUGU-GCACAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC --------((..((.(-(((((((((.....))))).))-)))..((((((((((((((((((..(((.....)))..........)))))))))))))))))).))..))... ( -30.00, z-score = -3.88, R) >droSim1.chr2R 14437799 104 - 19596830 --------GUAUGUAU-GUACGUGCGGCGGUCGCACUGU-GCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC --------((..((.(-(((((((((.....))))).))-)))..((((((((((((((((((..(((.....)))..........)))))))))))))))))).))..))... ( -30.30, z-score = -4.03, R) >droVir3.scaffold_12875 16778224 114 + 20611582 GUGUGUGUGCUUGUGGCUGAGGGGAUAUGAAUAUGUUGUCAUAAAGAAUGAAAAUUAAUUGAAAAUGCAUUCUGUAUGCCAUUGUCUUCAAUUAAUUUUCAUUUCAAUCAUUUC ............(((..(((((.(((((....))))).))......((((((((((((((((((((((((.....))).))))...))))))))))))))))))))..)))... ( -26.40, z-score = -1.71, R) >droGri2.scaffold_15112 2353926 106 - 5172618 GUGUGUGUGUGUGU-GCGGAGAGGAUAUGAAUAUGUUGUCAUAAAGAAUGAAAAUUAAUUGAAAAUGCAUUUUGCAUGCCAUUGUCUUCAAUUAAUUUUCAUUUCAA------- (((..(....)..)-))...((.(((((....))))).)).....(((((((((((((((((((((((((.....))).))))...))))))))))))))))))...------- ( -24.40, z-score = -1.27, R) >consensus ________GUAUGUAU_GUACGUGCGGCGGUCGCACUGU_GCAAAGAAUGAAAAUUAAUUGAAAUUGCAUUUUGCAUUCCAUUGUCUUCAAUUAAUUUUCAUUUCACUCACUUC .............................................((((((((((((((((((...(((...((.....)).))).)))))))))))))))))).......... (-16.95 = -16.95 + -0.00)
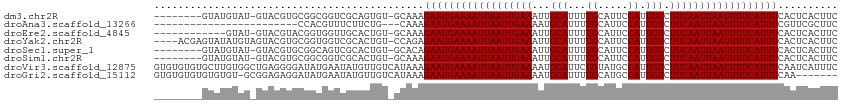

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:41 2011