| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,381,199 – 3,381,345 |
| Length | 146 |
| Max. P | 0.500000 |

| Location | 3,381,199 – 3,381,345 |
|---|---|
| Length | 146 |
| Sequences | 6 |
| Columns | 172 |
| Reading direction | reverse |
| Mean pairwise identity | 63.00 |
| Shannon entropy | 0.68941 |
| G+C content | 0.35903 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -11.34 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
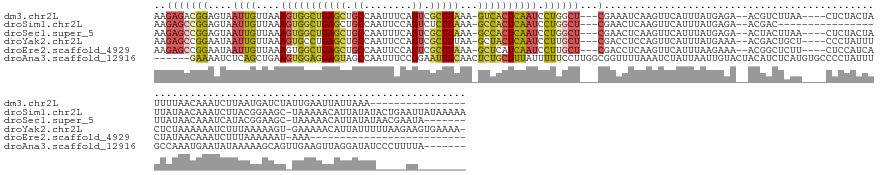
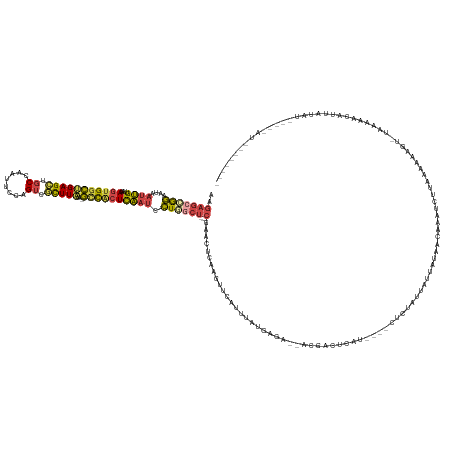
>dm3.chr2L 3381199 146 - 23011544 AAGAGACGGAGUAAUUGUUAAAGUGGCUGAGCUGCCAAUUUCAGUCGCUUAAA-GUCACUCAAUCCUGGCU---CGAAAUCAAGUUCAUUUAUGAGA--ACGUCUUAA----CUCUACUAUUUUAACAAAUCUUAAUGAUCUAUUGAAUUAUUAAA---------------- ..((((((((((.(((....(((((((((((........)))))))))))..)-)).))))........((---((((((.......)))).)))).--.))))))..----....................(((((((((....).)))))))).---------------- ( -31.00, z-score = -1.23, R) >droSim1.chr2L 3319874 149 - 22036055 AAGAGCCGGAGUAAUUGUUAAAGUGGCUGAGCUGCCAAUUCCAGUCUCUUAAA-GCCACUCAAUCCUGGCU---CGAACUCAAGUUCAUUUAUGAGA--ACGAC----------------UUAUAACAAAUCUUACGGAAGC-UAAAAACAUUAUAUACUGAAUUAUAAAAA ..(((((((.(..((((....(((((((((((((.......))).)))....)-)))))))))))))))))---)((((....))))..(((((((.--....)----------------))))))......((((....).-))).....(((((........)))))... ( -35.30, z-score = -2.50, R) >droSec1.super_5 1528209 154 - 5866729 AAGAGCCGGAGUAAUUGUUAAAGUGGCUGAGCUGCCAAUUUCAGUCGCUUAAA-GCCACUCAAUCCUGGCU---CGAACUCAAGUUCAUUUAUGAGA--ACUACUUAA----CUCUACUAUUAUAACAAAUCAUACGGAAGC-UAAAAACAUUAUAUAACGAAUA------- ..(((((((.(..((((....(((((((((((.((........)).))))..)-)))))))))))))))))---)....((.(((((........))--)))......----.........(((((.........(....).-........)))))....))...------- ( -32.67, z-score = -1.52, R) >droYak2.chr2L 3366083 160 - 22324452 AAGAGCCGGAAUAAUUGUUAAAGUGCCUGAGCUGCCAAUUCCAGUCGCUUUAA-GCUACUCAAUCCUUGCU---CGACCUCCAGUUCAUUUAUGAAA--ACGACUGCU----CCCUAUUUCUCUAAAAAAUCUUUAAAAAGU-GAAAAACAUUAUUUUUAAGAAGUGAAAA- ..((((.(((......(((((((((.(((............))).)))))).)-)).......)))..)))---)......(((((...........--..)))))..----.....((((.((......((((..((((((-((......)))))))))))))).)))).- ( -24.04, z-score = 0.46, R) >droEre2.scaffold_4929 3411102 135 - 26641161 AAGAGCCGGAAUAAUUGUUAAAGUGGCUGAGCUGCCAAUUCCAGUCGCUUAAA-GCUCAUCAAUCCUUGCU---CGACCUCAAGUUCAUUUAAGAAA--ACGGCUCUU----CUCCAUCACUAUAACAAAUCUUUAAAAAAU-AAA-------------------------- ((((((((............(((((((((............)))))))))...-...........((((..---......)))).............--.))))))))----..............................-...-------------------------- ( -24.70, z-score = -1.00, R) >droAna3.scaffold_12916 2836958 159 + 16180835 ------GAAAAUCUCAGCUGAAGUGGAGGAGUAGCCAAUUUCCGGAAUUUCAACUCUGCUUUAUUUUUCCUUGGCGGUUUUAAAUCUAUUAAUUGUACUACAUCUCAUGUGCCCCUAUUUGCCAAAUGAAUAUAAAAAGCAGUUGAAGUUAGGAUAUCCCUUUUA------- ------.............((((.(((((.....))....(((..((((((((((((..(((((.(((..((((((((.....)))........((((..........))))........)))))..))).))))).)).)))))))))).)))..)))))))..------- ( -33.60, z-score = -0.36, R) >consensus AAGAGCCGGAAUAAUUGUUAAAGUGGCUGAGCUGCCAAUUCCAGUCGCUUAAA_GCCACUCAAUCCUGGCU___CGAACUCAAGUUCAUUUAUGAGA__ACGACUCAU____CUCUAUUAUUAUAACAAAUCUUAAAAAAGU_UAAAAACAUUAUAU_____AU________ ...((((((....((((....(((((((((((.((........)).)))))...)))))))))).))))))..................................................................................................... (-11.34 = -11.35 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:41 2011