
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,766,870 – 15,766,968 |
| Length | 98 |
| Max. P | 0.514226 |

| Location | 15,766,870 – 15,766,968 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 62.79 |
| Shannon entropy | 0.72228 |
| G+C content | 0.56753 |
| Mean single sequence MFE | -32.49 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
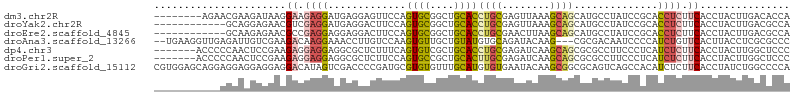
>dm3.chr2R 15766870 98 - 21146708 --------AGAACGAAGAUAAGGAAGAGGAUGAGGAGUUCCAGUGCGGCUGCACCUGCGAGUUAAAGCAGCAUGCCUAUCCGCACCUCUUCACCUACUUGACACCA --------........(.((((....(((.(((((((.....((((((..(((.((((........))))..)))....)))))))))))))))).)))).).... ( -31.60, z-score = -2.16, R) >droYak2.chr2R 7701881 94 + 21139217 ------------GCAGGAGAACGUCGAGGAUGAGGACUUCCAGUGCGGCUGCACCUGCGAGUUAAAGCAGCAUGCCUAUCCGCACCUCUUCACCUACUUGACGCCA ------------...((....(((((((..((((((......((((((..(((.((((........))))..)))....)))))).))))))....))))))))). ( -32.10, z-score = -1.43, R) >droEre2.scaffold_4845 9929427 94 - 22589142 ------------GCAAGAGAACGCCGAGGAGGAGGACUUCCAGUGCGGCUGCACCUGCGAACUUAAGCAGCAUGCCUAUCCGCACCUCUUCACCUACUUGACGCCA ------------.((((.(.....)(((((((.((....))..(((((..(((.((((........))))..)))....)))))))))))).....))))...... ( -29.90, z-score = -1.26, R) >droAna3.scaffold_13266 10325279 101 + 19884421 --UGAAGGUUGAGAUUGUCGAAGACAAGGAAACCUUGUCCAAGUGUUGCUGUAUGUGCAGAUACAAG---CGCGACAAUCCCCAUCUGUUCACUUACCUCGCGCCC --(((((((.(.(((((((...(((((((...)))))))...(((((.((((....)))).....))---)))))))))).).))))..))).............. ( -29.10, z-score = -2.06, R) >dp4.chr3 12894842 99 - 19779522 -------ACCCCCAACUCCGAAGAGGAGGAGGCGCUCUUUCAGUGUCGCUGCACCUGCGAGAUCAAGCAGCGCGCCUUCCCUCAUCUCUUCACCUACUUGGCUCCC -------....((((....((((((((((((((((.((((..((.((((.......)))).))..)).)).))))))..))))..))))))......))))..... ( -33.40, z-score = -1.76, R) >droPer1.super_2 307981 99 + 9036312 -------ACCCCCAACUCCGAAGAGGAGGAGGCGCUCUUCCAGUGCCGCUGCACUUGCGAGAUCAAGCAGCGCGCCUUCCCUCAUCUCUUCACCUACUUGGCUCCC -------....((((....((((((((((((((((......(((((....)))))(((........)))..))))))..))))..))))))......))))..... ( -33.40, z-score = -1.73, R) >droGri2.scaffold_15112 2339112 106 - 5172618 CGUGGAGCAGGAGGAGGAGGAGGACAUAGUCGACCCCGAUGCGUGUGUUUGCAUGUGUGAAUACAAGCGGCGCAGUCAGCCACAUCUCUUCACCUAUCUGGCCCCA ..(((.((((((((..(((((((.....((((....))))((.((((((..(....)..)))))).))(((.......)))...))))))).))).))).)).))) ( -37.90, z-score = -0.85, R) >consensus ___________AGAAAGACGAAGACGAGGAGGAGCACUUCCAGUGCGGCUGCACCUGCGAGUUCAAGCAGCGCGCCUAUCCGCAUCUCUUCACCUACUUGGCGCCA ......................((.((((.............((((....))))((((........))))..............)))).))............... ( -8.82 = -9.13 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:36 2011