| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,722,126 – 15,722,242 |
| Length | 116 |
| Max. P | 0.977748 |

| Location | 15,722,126 – 15,722,242 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.34618 |
| G+C content | 0.45565 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -18.51 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
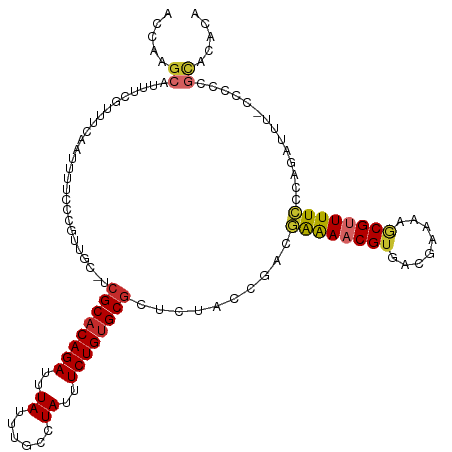
>dm3.chr2R 15722126 116 - 21146708 ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCAGAUUU-CCCCCGCACACA .....((.(((((((....(((((..((((.-.((((((((..((......))..))))))))......)))))))))...))))))).))..............-............ ( -26.10, z-score = -2.90, R) >droSim1.chr2R 14381208 116 - 19596830 ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCAGAUUU-CCCCCGCACACA .....((.(((((((....(((((..((((.-.((((((((..((......))..))))))))......)))))))))...))))))).))..............-............ ( -26.10, z-score = -2.90, R) >droSec1.super_1 13212318 116 - 14215200 ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCAGAUUU-CCCCCGCUCACA ....(((...................((((.-.((((((((..((......))..))))))))......))))((((((((........))))))))........-.....))).... ( -26.20, z-score = -2.90, R) >droYak2.chr2R 7655277 117 + 21139217 ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCAGUUUUGCCCCCGCUCACA .....((.(((((((....(((((..((((.-.((((((((..((......))..))))))))......)))))))))...))))))).)).........((...((....))..)). ( -26.60, z-score = -2.52, R) >droEre2.scaffold_4845 9884243 116 - 22589142 ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACAAAAACGUGACGAAAAGCGUUUUCCCAGAUUU-CCCCCGCACACA .....((.(((((((....((((..((..(.-.((((((((..((......))..))))))))..)...))..))))....))))))).))..............-............ ( -24.20, z-score = -2.63, R) >droAna3.scaffold_13266 10278832 116 + 19884421 ACCAAGCAGUUCGUUUCAAUUUUCCCGCCGC-UCGCUCAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCACAUUUUCCCCACCAGCC- .....((..((((((....(((((.((....-.(((.((((..((......))..)))).)))......))..)))))...))))))..))..........................- ( -20.80, z-score = -1.27, R) >dp4.chr3 12848912 114 - 19779522 ACCAAGCAUUUCAUUUCAUUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUGCCGAGAAGAACGUGACGAAAAACGAUUUUCC-CACUCGCCUAUGUAUG-- .....((...........((((((.((..((-.((((((((..((......))..))))))))....)))).)))))).(((..(((((....))))).-)))..)).........-- ( -24.00, z-score = -1.86, R) >droPer1.super_2 261104 114 + 9036312 GCCAAGCAUUUCAUUUCAUUUUUCCCGUUGC-UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUGCCGAGAAGAACGUGACGAAAAACGCUUUUCC-CAUUCGCCUAUGUAUG-- ..((.((((.........((((((.((..((-.((((((((..((......))..))))))))....)))).)))))).((((.(((((....))))).-...))))..)))).))-- ( -23.80, z-score = -1.35, R) >droWil1.scaffold_180701 1918882 100 - 3904529 ACCAAGCAUUUUAUUUCAAUUUUCUCGUUUUGUGGCACAGAUUUAUUUGCCUAUUUCUGUGCG----------AGGAACGUGACGAAUACCGUUUUUUUCCUGGCAGCUG-------- ....(((...........................(((((((..((......))..))))))).----------(((((...((((.....))))...)))))....))).-------- ( -23.30, z-score = -1.70, R) >consensus ACCAAGCAUUUCGUUUCAAUUUUCCCGUUGC_UCGCACAGAUUUAUUUGCCUAUUUCUGUGCGCUCUACCGACGAAAACGUGACGAAAAGCGUUUUCCCAGAUUU_CCCCCGCACACA .....((..........................((((((((..((......))..))))))))..........((((((((........))))))))..............))..... (-18.51 = -18.52 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:29 2011