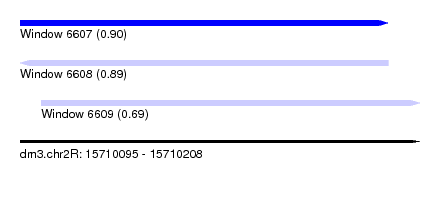
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,710,095 – 15,710,208 |
| Length | 113 |
| Max. P | 0.900586 |

| Location | 15,710,095 – 15,710,199 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
| Shannon entropy | 0.47935 |
| G+C content | 0.56094 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
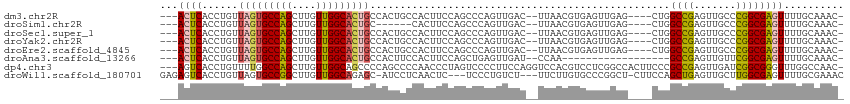
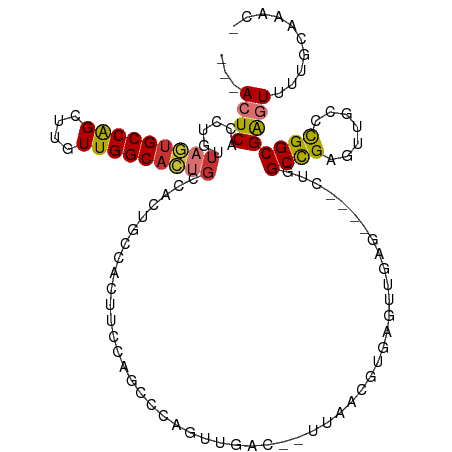
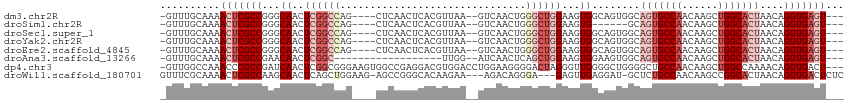
>dm3.chr2R 15710095 104 + 21146708 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUUGAC--UUAACGUGAGUUGAG----CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC- ---((((.......((((((((....))))))))(((...((.(((((((((.((..(.((--.....)).)..)).)----))))..)))).))..))))))).........- ( -37.70, z-score = -2.05, R) >droSim1.chr2R 14368842 98 + 19596830 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGC------CACUUCCAGCCCAGUUGAC--UUAACGUGAGUUGAG----CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC- ---((((.((((.(((((((((....))))))))).------))....((((.(.(((..(--(((((....))))))----..))).).))))...)).)))).........- ( -35.70, z-score = -2.17, R) >droSec1.super_1 13200355 104 + 14215200 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUUGAC--UUAACGUGAGUUGAG----CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC- ---((((.......((((((((....))))))))(((...((.(((((((((.((..(.((--.....)).)..)).)----))))..)))).))..))))))).........- ( -37.70, z-score = -2.05, R) >droYak2.chr2R 7643205 104 - 21139217 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUUGAC--UUAACGUGAGUUGAG----CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC- ---((((.......((((((((....))))))))(((...((.(((((((((.((..(.((--.....)).)..)).)----))))..)))).))..))))))).........- ( -37.70, z-score = -2.05, R) >droEre2.scaffold_4845 9872258 104 + 22589142 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUUGAC--UUAACGUGAGUUGAG----CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC- ---((((.......((((((((....))))))))(((...((.(((((((((.((..(.((--.....)).)..)).)----))))..)))).))..))))))).........- ( -37.70, z-score = -2.05, R) >droAna3.scaffold_13266 10266859 90 - 19884421 ---ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUUCCACUUCCAGCUGAGUUGAU--CCAA------------------GCCGAGUUGUUCGGCGAGUUUUGCAAAC- ---((((.((((.(((((((((....))))))))).))..........(((((..(((...--...)------------------))..)))))...)).)))).........- ( -26.70, z-score = -1.15, R) >dp4.chr3 12836032 110 + 19779522 ---AGUCACCUGUUUUGGCCAGCUUGUUGGCAGCCCCAGCCCCAACCCUAGUCCCCUUCCAGGUCCACGUCCUCGGCCACUUCCCGCCGAGUUGAUCGGCGGGUUUGGCCAAC- ---.((.(((((....((...(((.(((((..((....)).)))))...)))..))...)))))..))......(((((...((((((((.....))))))))..)))))...- ( -42.90, z-score = -3.14, R) >droWil1.scaffold_180701 1899011 106 + 3904529 GAGAGUCACCUGUUAGUGCCGGCUUGUUGGCAGAGC-AUCCUCAACUC---UCCCUGUCU---UUCUUGUGCCCGGCU-CUUCCAGCUGAGUUGCUUGGCGAGUUUUGCGAAAC ((((((....((((..((((((....)))))).)))-)......))))---)).......---......(((..((((-(..(((((......)).))).)))))..))).... ( -28.80, z-score = 0.78, R) >consensus ___ACUCACCUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUUGAC__UUAACGUGAGUUGAG____CUGGCCGAGUUGCCCGGCGAGUUUUGCAAAC_ ...((((......(((((((((....)))))))))..................................................((((.......)))))))).......... (-19.41 = -19.66 + 0.25)
| Location | 15,710,095 – 15,710,199 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.29 |
| Shannon entropy | 0.47935 |
| G+C content | 0.56094 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -18.94 |
| Energy contribution | -20.41 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15710095 104 - 21146708 -GUUUGCAAAACUCGCCGGGCAACUCGGCCAG----CUCAACUCACGUUAA--GUCAACUGGGCUGGAAGUGGCAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((..((.(((...((((----((((....((.....--))....)))))))).))).))..((..(((((((......)))))))..)))))))))--- ( -41.50, z-score = -2.14, R) >droSim1.chr2R 14368842 98 - 19596830 -GUUUGCAAAACUCGCCGGGCAACUCGGCCAG----CUCAACUCACGUUAA--GUCAACUGGGCUGGAAGUG------GCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((..((.(((...((((----((((....((.....--))....)))))))).))).------))(((((((......)))))))....)))))))--- ( -39.50, z-score = -2.71, R) >droSec1.super_1 13200355 104 - 14215200 -GUUUGCAAAACUCGCCGGGCAACUCGGCCAG----CUCAACUCACGUUAA--GUCAACUGGGCUGGAAGUGGCAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((..((.(((...((((----((((....((.....--))....)))))))).))).))..((..(((((((......)))))))..)))))))))--- ( -41.50, z-score = -2.14, R) >droYak2.chr2R 7643205 104 + 21139217 -GUUUGCAAAACUCGCCGGGCAACUCGGCCAG----CUCAACUCACGUUAA--GUCAACUGGGCUGGAAGUGGCAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((..((.(((...((((----((((....((.....--))....)))))))).))).))..((..(((((((......)))))))..)))))))))--- ( -41.50, z-score = -2.14, R) >droEre2.scaffold_4845 9872258 104 - 22589142 -GUUUGCAAAACUCGCCGGGCAACUCGGCCAG----CUCAACUCACGUUAA--GUCAACUGGGCUGGAAGUGGCAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((..((.(((...((((----((((....((.....--))....)))))))).))).))..((..(((((((......)))))))..)))))))))--- ( -41.50, z-score = -2.14, R) >droAna3.scaffold_13266 10266859 90 + 19884421 -GUUUGCAAAACUCGCCGAACAACUCGGC------------------UUGG--AUCAACUCAGCUGGAAGUGGAAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU--- -.........(((((((...((..(((((------------------(.((--.....)).))))))...))....((..(((((((......)))))))..)))))))))--- ( -31.50, z-score = -1.73, R) >dp4.chr3 12836032 110 - 19779522 -GUUGGCCAAACCCGCCGAUCAACUCGGCGGGAAGUGGCCGAGGACGUGGACCUGGAAGGGGACUAGGGUUGGGGCUGGGGCUGCCAACAAGCUGGCCAAAACAGGUGACU--- -.(((((((..((((((((.....))))))))...)))))))........(((((.....((.((((.((((((((....))).)))))...))))))....)))))....--- ( -52.60, z-score = -2.89, R) >droWil1.scaffold_180701 1899011 106 - 3904529 GUUUCGCAAAACUCGCCAAGCAACUCAGCUGGAAG-AGCCGGGCACAAGAA---AGACAGGGA---GAGUUGAGGAU-GCUCUGCCAACAAGCCGGCACUAACAGGUGACUCUC ((((.((.......)).))))...(((.(((..((-.(((((.(.(.....---.)...((.(---((((.......-))))).)).....)))))).))..))).)))..... ( -32.20, z-score = -0.43, R) >consensus _GUUUGCAAAACUCGCCGGGCAACUCGGCCAG____CUCAACUCACGUUAA__GUCAACUGGGCUGGAAGUGGCAGUGGCAGUGCCAACAAGCUGGCACUAACAGGUGAGU___ ..........(((((((.(((......)))..................................................(((((((......)))))))....)))))))... (-18.94 = -20.41 + 1.47)
| Location | 15,710,101 – 15,710,208 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Shannon entropy | 0.35591 |
| G+C content | 0.55511 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -24.22 |
| Energy contribution | -25.51 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
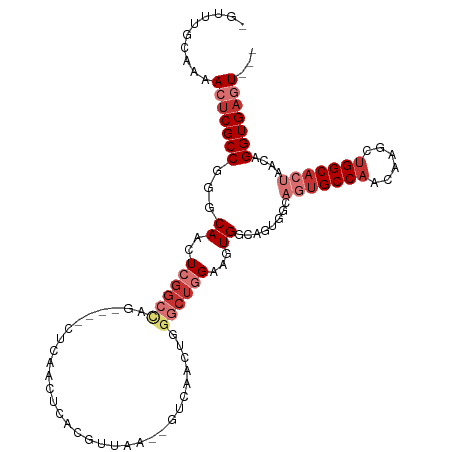
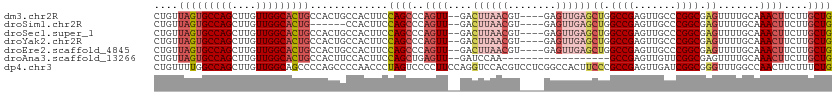
>dm3.chr2R 15710101 107 + 21146708 CUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUU--GACUUAACGU----GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..((.(((((((((.((..(--.((.....))----.)..)).)))))..)))).)).(((((((............))))))) ( -37.20, z-score = -1.48, R) >droSim1.chr2R 14368848 101 + 19596830 CUGUUAGUGCCAGCUUGUUGGCACUG------CCACUUCCAGCCCAGUU--GACUUAACGU----GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....)))))))))------.))....((((..((((--..((((((..----..))))))((.((((.......)))).)).......))))....)))) ( -35.70, z-score = -1.70, R) >droSec1.super_1 13200361 107 + 14215200 CUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUU--GACUUAACGU----GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..((.(((((((((.((..(--.((.....))----.)..)).)))))..)))).)).(((((((............))))))) ( -37.20, z-score = -1.48, R) >droYak2.chr2R 7643211 107 - 21139217 CUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUU--GACUUAACGU----GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..((.(((((((((.((..(--.((.....))----.)..)).)))))..)))).)).(((((((............))))))) ( -37.20, z-score = -1.48, R) >droEre2.scaffold_4845 9872264 107 + 22589142 CUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUU--GACUUAACGU----GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..((.(((((((((.((..(--.((.....))----.)..)).)))))..)))).)).(((((((............))))))) ( -37.20, z-score = -1.48, R) >droAna3.scaffold_13266 10266865 93 - 19884421 CUGUUAGUGCCAGCUUGUUGGCACUGCCACUUCCACUUCCAGCUGAGUU--GAUCCAA------------------GCCGAGUUGUUCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..........((((.(((((--((..(..------------------((((((...))))))..)..))...)))))...)))) ( -28.40, z-score = -1.13, R) >dp4.chr3 12836038 113 + 19779522 CUGUUUUGGCCAGCUUGUUGGCAGCCCCAGCCCCAACCCUAGUCCCCUUCCAGGUCCACGUCCUCGGCCACUUCCCGCCGAGUUGAUCGGCGGGUUUGGCCAACUUCUUUCUG (((....((...(((.(((((..((....)).)))))...)))..))...)))............(((((...((((((((.....))))))))..)))))............ ( -38.60, z-score = -2.07, R) >consensus CUGUUAGUGCCAGCUUGUUGGCACUGCCACUGCCACUUCCAGCCCAGUU__GACUUAACGU____GAGUUGAGCUGGCCGAGUUGCCCGGCGAGUUUUGCAAACUUCUUGCUG .((.(((((((((....))))))))).))..........((((...((....)).............((.(((((.((((.......)))).))))).)).........)))) (-24.22 = -25.51 + 1.29)
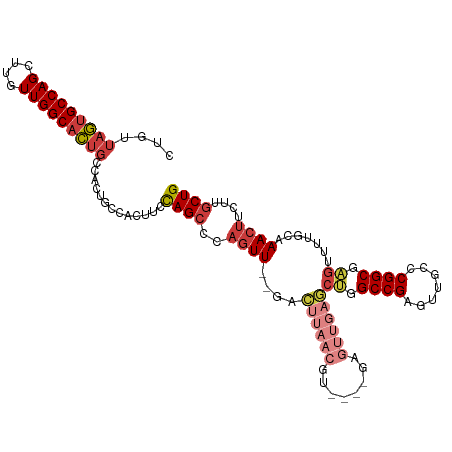
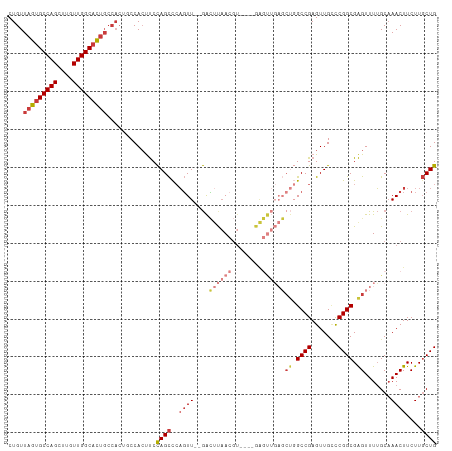
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:25 2011