
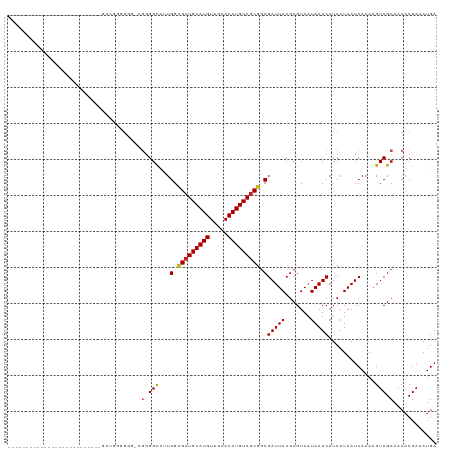
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,694,673 – 15,694,788 |
| Length | 115 |
| Max. P | 0.950525 |

| Location | 15,694,673 – 15,694,774 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Shannon entropy | 0.36038 |
| G+C content | 0.48468 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.29 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15694673 101 + 21146708 -----------------UGGAUGGGUGAUGGGGGG-CGGGGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA -----------------..(((..(((((((...(-((.(..((((.(((((((((.....))))))))).))))..).))).....))))))).))).....(((.....)))..... ( -36.60, z-score = -3.89, R) >droSim1.chr2R 14353393 101 + 19596830 -----------------UGGAUGGGUGAUGGGGGG-CGGGGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA -----------------..(((..(((((((...(-((.(..((((.(((((((((.....))))))))).))))..).))).....))))))).))).....(((.....)))..... ( -36.60, z-score = -3.89, R) >droSec1.super_1 13184936 102 + 14215200 -----------------UGGUUGGGUGAUGGGGGGGCGGGGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA -----------------((((((((((((...(..(((.(..((((.(((((((((.....))))))))).))))..).)))..).)))))).))))))....(((.....)))..... ( -36.80, z-score = -3.83, R) >droEre2.scaffold_4845 9857161 101 + 22589142 -----------------AAGAUGGUAGAUGGGGGG-CGGGGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA -----------------..(((.((.(((((...(-((.(..((((.(((((((((.....))))))))).))))..).))).....))))))).))).....(((.....)))..... ( -33.20, z-score = -3.18, R) >droAna3.scaffold_13266 10251989 92 - 19884421 ---------------------------AGCAGGAGUGGGGGUACUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA ---------------------------..((...(((..((.((((.(((((((((.....))))))))).)(((((.......)))))............)))).)..)))....)). ( -25.10, z-score = -0.87, R) >dp4.chr3 12819351 92 + 19779522 --------------------------GAUAGAGGA-GGGGGCCUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA --------------------------(((.((..(-..(.((.(((.(((((((((.....))))))))).)))....)).)..)..)))))....(((....(((.....)))..))) ( -24.40, z-score = -0.85, R) >droPer1.super_2 231513 92 - 9036312 --------------------------GAUAGUGGA-GGGGGCCUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA --------------------------....(((..-.(.(((...(.(((((((((.....))))))))).)(((((.......))))).............))).)..)))....... ( -26.90, z-score = -1.35, R) >droWil1.scaffold_180701 1869670 80 + 3904529 ---------------------------------------GGUAGUGGUGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA ---------------------------------------(((.(((((((((((((.....)))))))).(((....))).........))))).))).....(((.....)))..... ( -21.30, z-score = -0.95, R) >droVir3.scaffold_12875 16652900 83 - 20611582 ------------------------------------UGGGGCGAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA ------------------------------------((..((((((.(((((((((.....))))))))).)(((((.......))))).............)))))..))........ ( -27.90, z-score = -2.54, R) >droMoj3.scaffold_6496 21093958 89 + 26866924 ------------------------------UGGCAAUGGGGCGAUGGCGCUGCAUGUAGCACAUGUAUCAACGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA ------------------------------......((..(((((...(.((((((.....)))))).)...(((((.......))))).............)))))..))........ ( -17.40, z-score = 1.06, R) >droGri2.scaffold_15112 2237295 119 + 5172618 GGAAGGGGGGGGGGGUGCGUUGGGACUGGGGCGGGUGAUGGCAAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA ...............((.((((.((((.((...((((((.((.....(((((((((.....)))))))))(((....)))))....))))))....))...)))).)))).))...... ( -38.40, z-score = -2.90, R) >consensus __________________________GAUGGGGGG_CGGGGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGA .....................................(.(((...(.(((((((((.....))))))))).)(((((.......))))).............))).)............ (-20.15 = -20.29 + 0.14)


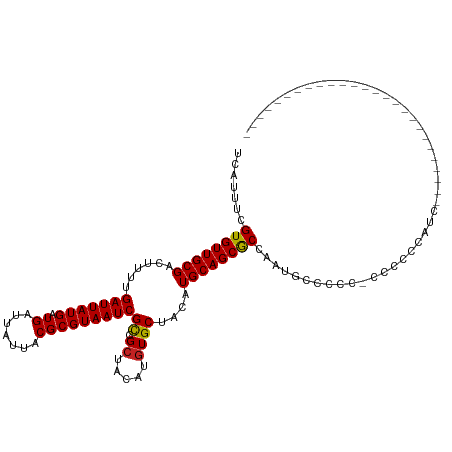
| Location | 15,694,673 – 15,694,774 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.80 |
| Shannon entropy | 0.36038 |
| G+C content | 0.48468 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.29 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15694673 101 - 21146708 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAUGCCCCG-CCCCCCAUCACCCAUCCA----------------- .......(((..(((......(((((((.((.......)))))))))(.((((.((((.....)))).)))).)..)))..))-).................----------------- ( -19.60, z-score = -0.70, R) >droSim1.chr2R 14353393 101 - 19596830 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAUGCCCCG-CCCCCCAUCACCCAUCCA----------------- .......(((..(((......(((((((.((.......)))))))))(.((((.((((.....)))).)))).)..)))..))-).................----------------- ( -19.60, z-score = -0.70, R) >droSec1.super_1 13184936 102 - 14215200 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAUGCCCCGCCCCCCCAUCACCCAACCA----------------- .......(((..(((......(((((((.((.......)))))))))(.((((.((((.....)))).)))).)..)))..)))..................----------------- ( -19.60, z-score = -0.93, R) >droEre2.scaffold_4845 9857161 101 - 22589142 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAUGCCCCG-CCCCCCAUCUACCAUCUU----------------- .......(((..(((......(((((((.((.......)))))))))(.((((.((((.....)))).)))).)..)))..))-).................----------------- ( -19.60, z-score = -0.84, R) >droAna3.scaffold_13266 10251989 92 + 19884421 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAGUACCCCCACUCCUGCU--------------------------- .......(((.....(((...(((((((.((.......)))))))))(.((((.((((.....)))).)))).)))).....))).......--------------------------- ( -18.80, z-score = -0.40, R) >dp4.chr3 12819351 92 - 19779522 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAGGCCCCC-UCCUCUAUC-------------------------- .......((((((((......(((((((.((.......)))))))))((.((.....))))....))))))))..(((.....-.))).....-------------------------- ( -18.30, z-score = -0.10, R) >droPer1.super_2 231513 92 + 9036312 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAGGCCCCC-UCCACUAUC-------------------------- .......((((((((.....(((((.....))))).....))))).))).(((.((((.....)))).)))((....))....-.........-------------------------- ( -18.20, z-score = 0.03, R) >droWil1.scaffold_180701 1869670 80 - 3904529 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCACCACUACC--------------------------------------- .......((((((((......(((((((.((.......)))))))))((.((.....))))....)))))))).......--------------------------------------- ( -18.30, z-score = -1.18, R) >droVir3.scaffold_12875 16652900 83 + 20611582 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAUCGCCCCA------------------------------------ ............((((.....(((((((.((.......)))))))))(.((((.((((.....)))).)))).).))))....------------------------------------ ( -20.20, z-score = -0.73, R) >droMoj3.scaffold_6496 21093958 89 - 26866924 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGUUGAUACAUGUGCUACAUGCAGCGCCAUCGCCCCAUUGCCA------------------------------ .......(((((((.............(((((((((.....))))))))).....(((.....))))))))))................------------------------------ ( -16.80, z-score = 0.91, R) >droGri2.scaffold_15112 2237295 119 - 5172618 UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAUUGCCAUCACCCGCCCCAGUCCCAACGCACCCCCCCCCCCUUCC .......((((((.((((........(((((........(((....)))((((.((((.....)))).)))).......)))))........)))).))))))................ ( -29.29, z-score = -4.10, R) >consensus UCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAAUGCCCCC_CCCCCCAUC__________________________ .......((((((((......(((((((.((.......)))))))))((.((.....))))....)))))))).............................................. (-17.36 = -17.29 + -0.07)

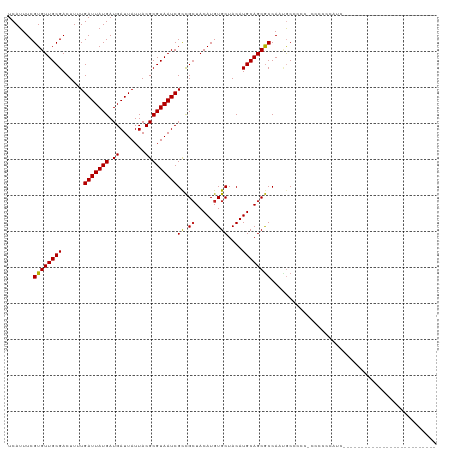
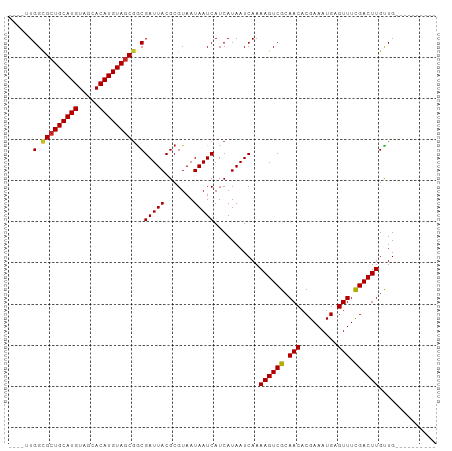
| Location | 15,694,698 – 15,694,788 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 93.41 |
| Shannon entropy | 0.13504 |
| G+C content | 0.43559 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15694698 90 + 21146708 ----UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGCUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -26.30, z-score = -1.38, R) >droSim1.chr2R 14353418 90 + 19596830 ----UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGCUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -26.30, z-score = -1.38, R) >droSec1.super_1 13184962 90 + 14215200 ----UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGCUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -26.30, z-score = -1.38, R) >droYak2.chr2R 7627212 90 - 21139217 ----UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGCUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -26.30, z-score = -1.38, R) >droEre2.scaffold_4845 9857186 90 + 22589142 ----UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -26.30, z-score = -1.56, R) >droAna3.scaffold_13266 10252005 100 - 19884421 ----CUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUGUGAGCUGCCG ----..(((((((((((.....))))))))((((((((.......))))).(((((((...((((((.(((.........))).))))))))))))))))))). ( -36.00, z-score = -2.90, R) >droWil1.scaffold_180701 1869675 89 + 3904529 -----UGGUGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG---------- -----.(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -24.20, z-score = -0.96, R) >droVir3.scaffold_12875 16652907 90 - 20611582 ----AUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUUGACUUGUUG---------- ----..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).))))))....---------- ( -24.70, z-score = -0.95, R) >droMoj3.scaffold_6496 21093971 95 + 26866924 ----AUGGCGCUGCAUGUAGCACAUGUAUCAACGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUUGACUUGUUGUUGCU----- ----..((((.((((((.....)))))).)...(((((.......))))).............)))((((((....(..(((....)))..))))))).----- ( -20.20, z-score = 0.55, R) >droGri2.scaffold_15112 2237334 94 + 5172618 GGCAAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUUGACUUGUUG---------- (((((.(.(((((((((.....))))))))).)(((((.......))))).......((((((.(((..........))).)))))).))))).---------- ( -25.40, z-score = -0.72, R) >consensus ____UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG__________ ......(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).)))))).............. (-24.59 = -24.50 + -0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:22 2011