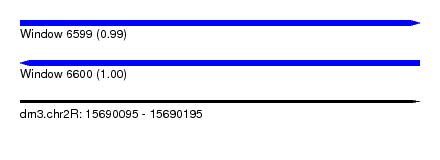
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,690,095 – 15,690,195 |
| Length | 100 |
| Max. P | 0.996580 |

| Location | 15,690,095 – 15,690,195 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.26 |
| Shannon entropy | 0.26287 |
| G+C content | 0.50607 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15690095 100 + 21146708 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA-CAAAUUUUUUGUUAAUUUUUUGUU-- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))..((-(((.....)))))...........-- ( -34.00, z-score = -2.41, R) >droSim1.chr2R 14348794 100 + 19596830 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA-CAACUUUUUUGUUAUAUUUCUGUU-- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((((-.(((......)))...))))....-- ( -34.00, z-score = -2.07, R) >droSec1.super_1 13180370 100 + 14215200 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA-CAACUUUUUUGUUAAUUUUUUGUU-- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))..((-(((.....)))))...........-- ( -33.90, z-score = -2.16, R) >droYak2.chr2R 7622336 101 - 21139217 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA-CAACUUUUUUGUUAAUCAUAUUUUU- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((((-(((.....)))))..))........- ( -34.00, z-score = -2.54, R) >droEre2.scaffold_4845 9852305 92 + 22589142 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAU--AACUUUUUUUUUGUU--------- (((((((..((((........))))....((((((.......))))))(((((.......)))))))))))).....--...............--------- ( -32.70, z-score = -2.14, R) >droAna3.scaffold_13266 10247186 95 - 19884421 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAUAUCUUUUUUGUCUAUCG-------- ((((.....((((........)))).(((((.......)))))))))((((((.......)))))).((((((((((.....))))))))))...-------- ( -32.90, z-score = -1.97, R) >dp4.chr3 12815156 103 + 19779522 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAACAAACUUUUUGUUUCUUCUUUUCCUG (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))(((((.((((.....))))....)))))... ( -34.60, z-score = -1.89, R) >droPer1.super_2 227225 103 - 9036312 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAACAAACUUUUUGUUUCUUCUUUUCCUG (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))(((((.((((.....))))....)))))... ( -34.60, z-score = -1.89, R) >droVir3.scaffold_12875 16646641 95 - 20611582 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA-CAAAUUUUUUGG-AUUUUAU------ (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))....-............-.......------ ( -32.70, z-score = -1.89, R) >droGri2.scaffold_15112 2229964 98 + 5172618 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAUAGAAUUUUUUGCUUUAAUUUC----- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))(((((.(((........))).)))))----- ( -33.50, z-score = -2.04, R) >anoGam1.chr3R 35066376 100 - 53272125 GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGCAA-CGAUUCUUUUGCUAUUUUGCUUCA-- (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))((((-........))))............-- ( -35.20, z-score = -1.46, R) >consensus GCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA_CAAAUUUUUUGUUAAUUUUUU_____ (((((((..((((........))))....((((((.......))))))(((((.......))))))))))))............................... (-32.70 = -32.70 + -0.00)

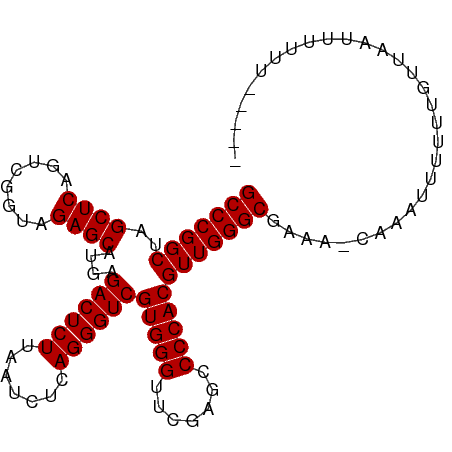
| Location | 15,690,095 – 15,690,195 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.26 |
| Shannon entropy | 0.26287 |
| G+C content | 0.50607 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15690095 100 - 21146708 --AACAAAAAAUUAACAAAAAAUUUG-UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC --...........(((((.....)))-))..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -30.40, z-score = -3.14, R) >droSim1.chr2R 14348794 100 - 19596830 --AACAGAAAUAUAACAAAAAAGUUG-UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC --....((((((..((......))))-))))((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -32.80, z-score = -3.26, R) >droSec1.super_1 13180370 100 - 14215200 --AACAAAAAAUUAACAAAAAAGUUG-UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC --...........(((((.....)))-))..((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -30.40, z-score = -2.72, R) >droYak2.chr2R 7622336 101 + 21139217 -AAAAAUAUGAUUAACAAAAAAGUUG-UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC -........((.((((......))))-..))((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -31.40, z-score = -2.98, R) >droEre2.scaffold_4845 9852305 92 - 22589142 ---------AACAAAAAAAAAGUU--AUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC ---------...............--.....((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -29.80, z-score = -2.92, R) >droAna3.scaffold_13266 10247186 95 + 19884421 --------CGAUAGACAAAAAAGAUAUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC --------.......................((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -29.80, z-score = -2.53, R) >dp4.chr3 12815156 103 - 19779522 CAGGAAAAGAAGAAACAAAAAGUUUGUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC .......((((.((((.....)))).)))).((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -32.80, z-score = -2.03, R) >droPer1.super_2 227225 103 + 9036312 CAGGAAAAGAAGAAACAAAAAGUUUGUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC .......((((.((((.....)))).)))).((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -32.80, z-score = -2.03, R) >droVir3.scaffold_12875 16646641 95 + 20611582 ------AUAAAAU-CCAAAAAAUUUG-UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC ------.......-............-....((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -29.80, z-score = -2.82, R) >droGri2.scaffold_15112 2229964 98 - 5172618 -----GAAAUUAAAGCAAAAAAUUCUAUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC -----(((((..((........))..)))))((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -31.70, z-score = -3.24, R) >anoGam1.chr3R 35066376 100 + 53272125 --UGAAGCAAAAUAGCAAAAGAAUCG-UUGCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC --....((((.....(....).....-))))((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) ( -33.00, z-score = -2.30, R) >consensus _____AAAAAAAUAACAAAAAAUUUG_UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGC ...............................((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))) (-29.80 = -29.80 + 0.00)
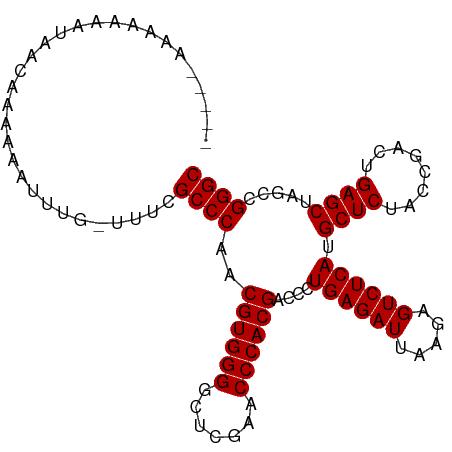
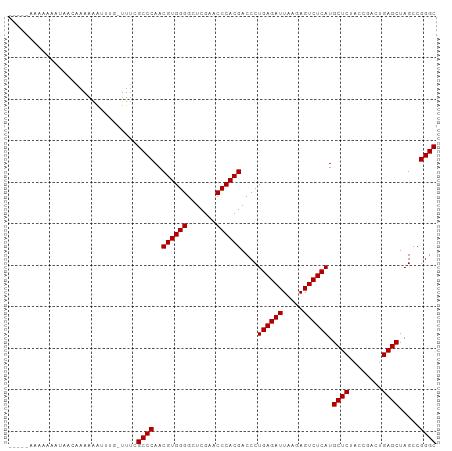
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:18 2011