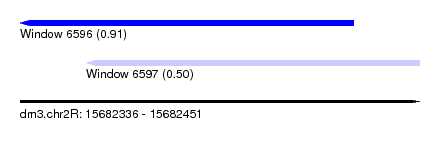
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,682,336 – 15,682,451 |
| Length | 115 |
| Max. P | 0.914421 |

| Location | 15,682,336 – 15,682,432 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.58082 |
| G+C content | 0.45819 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -9.22 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
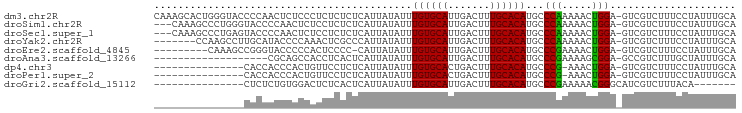
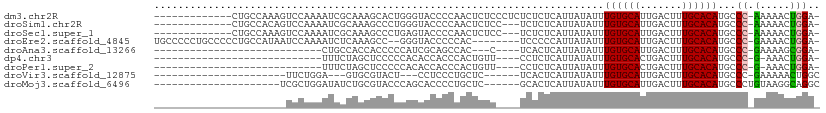
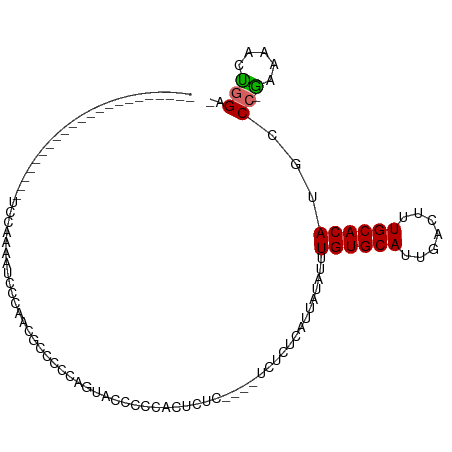
>dm3.chr2R 15682336 96 - 21146708 CAAAGCACUGGGUACCCCAACUCUCCCUCUCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCAAAAACUGGA-GUCGUCUUUCCUAUUUGCA ....(((.((((((.............................((((((.......))))))))))))......(((-(......))))....))). ( -18.85, z-score = -1.55, R) >droSim1.chr2R 14340284 93 - 19596830 ---CAAAGCCCUGGGUACCCCAACUCUCCUCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCAAAAACUGGA-GUCGUCUUUCCUAUUUGCA ---....((..((((((..........................((((((.......)))))))))))).(((..(((-(......))))..))))). ( -17.66, z-score = -1.16, R) >droSec1.super_1 13172431 93 - 14215200 ---CAAAGCCCUGAGUACCCCAACUCUCCUCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCAAAAACUGGA-GUCGUCUUUCCUAUUUGCA ---....((...((((......)))).................((((((.......)))))).......(((..(((-(......))))..))))). ( -14.90, z-score = -1.03, R) >droYak2.chr2R 7614036 89 + 21139217 -------CCAAGCCUUGCAUACCCCAAACUCGCCCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCAAAAACUGGA-GUCGUCUUUCCUAUUUGCA -------........((((........................((((((.......))))))............(((-(......))))....)))) ( -13.60, z-score = -0.37, R) >droEre2.scaffold_4845 9844226 86 - 22589142 ---------CAAAGCCGGGUACCCCCACUCCCC-CAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCGAAAACUGGA-GUCGUCUUUCCUAUUUGCA ---------....((((((((............-.........((((((.......)))))))))))).(((..(((-(......))))..))))). ( -19.19, z-score = -1.61, R) >droAna3.scaffold_13266 10238444 77 + 19884421 -------------------CGCAGCCACCUCACUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCGAAAAGCGGA-GCCGUCUUUGCUAUUUGCA -------------------.((((...................((((((.......)))))).........((((((-(....)))))))..)))). ( -17.30, z-score = -0.59, R) >dp4.chr3 12807140 80 - 19779522 ---------------CACCACCCACUGUUCCUCUCAUUAUAUUUGUGCACUGACUUUGCACAUGCCCG-AAACUGGA-GUCGUCUUUCCUAUUUGCA ---------------............................((((((.......))))))(((...-(((..(((-(......))))..)))))) ( -11.00, z-score = -0.18, R) >droPer1.super_2 218916 80 + 9036312 ---------------CACCACCCACUGUUCCUCUCAUUAUAUUUGUGCACUGACUUUGCACAUGCCCG-AAACUGGA-GUCGUCUUUCCUAUUUGCA ---------------............................((((((.......))))))(((...-(((..(((-(......))))..)))))) ( -11.00, z-score = -0.18, R) >droGri2.scaffold_15112 2217398 75 - 5172618 ---------------CUCUCUGUGGACUCUCACUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCGAAAAACGGGCAUCGUCUUUACA------- ---------------......((((((................((((((.......))))))((((((.....))))))..))))..)).------- ( -21.50, z-score = -4.00, R) >consensus _______________CACCCCCCCCCACCUCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCGAAAACUGGA_GUCGUCUUUCCUAUUUGCA ...........................................((((((.......))))))...(((.....)))..................... ( -9.22 = -8.80 + -0.42)
| Location | 15,682,355 – 15,682,451 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.63657 |
| G+C content | 0.48961 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -7.20 |
| Energy contribution | -7.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 15682355 96 - 21146708 -------------CUGCCAAAGUCCAAAAUCGCAAAGCACUGGGUACCCCAACUCUCCCUCUCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-AAAAACUGGA- -------------.........((((.....((...))..((((((.............................((((((.......)))))))))))-).....))))- ( -15.65, z-score = -0.17, R) >droSim1.chr2R 14340303 93 - 19596830 -------------CUGCCACAGUCCAAAAUCGCAAAGCCCUGGGUACCCCAACUCUCC---UCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-AAAAACUGGA- -------------......((((........(((......((((...)))).......---..............((((((.......)))))))))..-....))))..- ( -15.94, z-score = -0.22, R) >droSec1.super_1 13172450 93 - 14215200 -------------CUGCCAAAGUCCAAAAUCGCAAAGCCCUGAGUACCCCAACUCUCC---UCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-AAAAACUGGA- -------------.(((.((((((...(((((((((....((((..............---...)))).....)))))).)))))))))))).....((-(.....))).- ( -16.63, z-score = -1.71, R) >droEre2.scaffold_4845 9844245 99 - 22589142 UGCCCCCUGCCCCCUGCCAUAAUCCAAAAUCUCAAAGCC--GGGUACCCCCAC--------UCCCCCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-GAAAACUGGA- .....((.((.....)).....................(--(((((.......--------..............((((((.......)))))))))))-)......)).- ( -14.99, z-score = -0.00, R) >droAna3.scaffold_13266 10238463 74 + 19884421 ----------------------------CUGCCACCACCCCCAUCGCAGCCAC---C----UCACUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-GAAAAGCGGA- ----------------------------((((.............))))....---.----..............((((((.......))))))...((-(.....))).- ( -12.32, z-score = -0.52, R) >dp4.chr3 12807159 76 - 19779522 ----------------------------UUUCUAGCUCCCCCACACCACCCACUGUU----CCUCUCAUUAUAUUUGUGCACUGACUUUGCACAUGCCC-G-AAACUGGA- ----------------------------..(((((.......(((........))).----..............((((((.......)))))).....-.-...)))))- ( -8.90, z-score = -0.64, R) >droPer1.super_2 218935 76 + 9036312 ----------------------------UUUCUAGCUCCCCCACACCACCCACUGUU----CCUCUCAUUAUAUUUGUGCACUGACUUUGCACAUGCCC-G-AAACUGGA- ----------------------------..(((((.......(((........))).----..............((((((.......)))))).....-.-...)))))- ( -8.90, z-score = -0.64, R) >droVir3.scaffold_12875 16633259 76 + 20611582 ----------------------UUCUGGA---GUGCGUACU---CCUCCCUGCUC------UCACUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC-GAAAAACUGGC ----------------------.....((---(((.(..(.---.......)..)------.)))))........((((((.......)))))).(((.-(.....).))) ( -15.90, z-score = -1.30, R) >droMoj3.scaffold_6496 21069958 84 - 26866924 ---------------------UCGCUGGAUAUCUGCGUACCCAGCACCCCUGCUC------GCACUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCCUGUAAGGCAGGC ---------------------..(((((.(((....))).)))))...((((((.------((((...........)))).......(((((.......))))))))))). ( -24.40, z-score = -1.19, R) >consensus ______________________UCCAAAAUCCCAACGCCCCCAGUACCCCCACUCUC____UCUCUCAUUAUAUUUGUGCAUUGACUUUGCACAUGCCC_GAAAACUGGA_ ...........................................................................((((((.......))))))................. ( -7.20 = -7.20 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:15 2011