
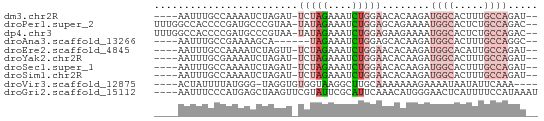
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,676,022 – 15,676,079 |
| Length | 57 |
| Max. P | 0.725564 |

| Location | 15,676,022 – 15,676,079 |
|---|---|
| Length | 57 |
| Sequences | 10 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 61.92 |
| Shannon entropy | 0.77577 |
| G+C content | 0.39446 |
| Mean single sequence MFE | -11.74 |
| Consensus MFE | -3.55 |
| Energy contribution | -4.61 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
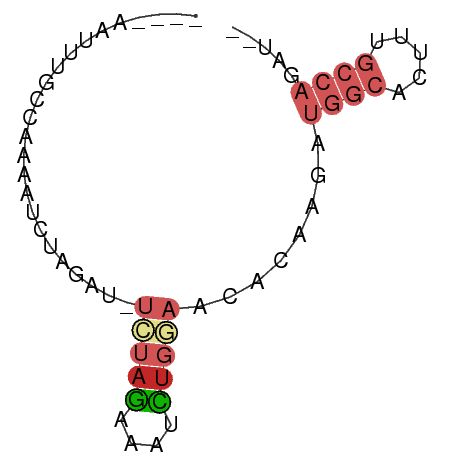
>dm3.chr2R 15676022 57 - 21146708 ----AAUUUGCCAAAAUCUAGAU-UCUAGAAAUCUGGAACACAAGAUGGCACUUUGCCAGAU-- ----............(((((((-(.....))))))))........((((.....))))...-- ( -12.80, z-score = -2.02, R) >droPer1.super_2 212721 61 + 9036312 UUUGGCCACCCCGAUGCCCGUAA-UAUAGAAAUCUGGAGCAGAAAAUGGCACUCUGCCAGAC-- .((((.....))))((((((...-..........))).))).....((((.....))))...-- ( -10.82, z-score = 0.19, R) >dp4.chr3 12800950 61 - 19779522 UUUGGCCACCCCGAUGCCCGUAA-UAUAGAAAUCUGGAGAAGAAAAUGGCACUCUGCCAGAC-- ((((((......(((((((....-....)...(((.....)))....)))).)).)))))).-- ( -9.60, z-score = 0.21, R) >droAna3.scaffold_13266 10232212 52 + 19884421 ----AAUUUGCCGAAAAGCA------UAGAAAUCUGGAGCACAAGAUGGCACUUUGCCAGGC-- ----.....(((.....((.------(((....)))..))......((((.....)))))))-- ( -12.70, z-score = -1.04, R) >droEre2.scaffold_4845 9837782 57 - 22589142 ----AAUUUGCCAAAAUCUAGUU-UCUAGAAAUCUGGAACACAAGAUGGCACAUUGCCAGAU-- ----(((.(((((...(((.(((-(((((....)))))).)).)))))))).))).......-- ( -14.90, z-score = -2.76, R) >droYak2.chr2R 7607327 57 + 21139217 ----AAUUUGCGAAAAUCUAGAU-UCUAGAAAUCUGGAACACAAGAUGGCACUUUGCCAGAU-- ----............(((((((-(.....))))))))........((((.....))))...-- ( -12.80, z-score = -2.12, R) >droSec1.super_1 13166119 57 - 14215200 ----AAUUUGCCAAAAUCUAGAU-UCUAGAAAUCUGGAACACAAGAUGGCACUUUGCCAGAU-- ----............(((((((-(.....))))))))........((((.....))))...-- ( -12.80, z-score = -2.02, R) >droSim1.chr2R 14334044 57 - 19596830 ----AAUUUGCCAAAAUCUAGAU-UCUAGAAAUCUGGAACACAAGAUGGCACUUUGCCAGAU-- ----............(((((((-(.....))))))))........((((.....))))...-- ( -12.80, z-score = -2.02, R) >droVir3.scaffold_12875 16623000 55 + 20611582 ----ACUAUUUUAUGGG-UAGGUGUGGUAAGGCUUGCAAAAAAAGAAAAUAAUAUUCAAA---- ----..((((((....(-(((((.(....).))))))........)))))).........---- ( -7.20, z-score = -0.78, R) >droGri2.scaffold_15112 2208944 60 - 5172618 ----AAUUUCCCAUGAGCUAAGUUCGUAUUCGCAUUCAAACAUGGGAACUCAUUUUCCAUAAAU ----...((((((((.....(((.((....)).)))....))))))))................ ( -11.00, z-score = -2.26, R) >consensus ____AAUUUGCCAAAAUCUAGAU_UCUAGAAAUCUGGAACACAAGAUGGCACUUUGCCAGAU__ ........................(((((....)))))........((((.....))))..... ( -3.55 = -4.61 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:14 2011