
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,366,393 – 3,366,502 |
| Length | 109 |
| Max. P | 0.549759 |

| Location | 3,366,393 – 3,366,502 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.41 |
| Shannon entropy | 0.61379 |
| G+C content | 0.45379 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -8.43 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3366393 109 - 23011544 UACACGCCCCGA-AAAAGGUGUGAAAUUGGUGGAGCCAAGUGUUCGAAUCAAAUUAA--AGGCACUCGAAGCGACGAUCA--------AUCGAUCAGGCGAUUGACAGAUAGAAAACUGC ..((((((....-....))))))....(((.....)))(((.(((..(((.......--..((.......))...(.(((--------((((......)))))))).))).))).))).. ( -24.70, z-score = -0.39, R) >droSim1.chr2L 3304525 109 - 22036055 CACACGCCCCGA-AAAAGGUGUGAAAUUGGUGGAGCCAAGUGUUCGAAUCAAAUUAA--AGCCACUCGAAGCGACGAUCA--------AUCGAUCAGCCGAUUGACAGAUAGAAAGCUGC ..((((((....-....))))))...(((((((.......((.......))......--..)))).)))(((....(((.--------.((((((....))))))..))).....))).. ( -25.46, z-score = -0.69, R) >droSec1.super_5 1513492 109 - 5866729 CACACGCCCCGA-AAAAGGUGUGAAAUUGGUAGAGCCAAGUGUUCGAAUCAAAUUAA--AGCCACUCGAAGCGACGAUCA--------AUCGAUCAGCCGAUUGACAGAUAGAAAGUUGC ..((((((....-....))))))...((((.....))))...(((((..........--......)))))(((((.(((.--------.((((((....))))))..))).....))))) ( -26.29, z-score = -1.79, R) >droYak2.chr2L 3351207 109 - 22324452 CACACGCCCCGA-AAAAGGUGUGAAAUUGGUGGAGCCAAGUGUUCGAAUCAAAUUAG--AGGCACUCGAAGCGGCGAUCA--------AUCGAUCAGGCGAUUGACAGAUAGAAAGCCGC ..((((((....-....))))))...((((.....))))((((((...........)--).)))).....(((((.(((.--------.((((((....))))))..))).....))))) ( -31.40, z-score = -1.52, R) >droEre2.scaffold_4929 3396442 100 - 26641161 ---------CGA-AAAAGGUGUGAAAUUGGUGGAGCCAAGUGUUCGAAUCAAAUUAA--AGGCACUCGAAGCGGCGAUCA--------AUCGAUCAGGCGAUUGACAGAUUGAAAGCUGC ---------...-....(((((.((.(((((.((((.....))))..))))).))..--..)))))....(((((..(((--------(((..((((....))))..))))))..))))) ( -27.40, z-score = -1.56, R) >droAna3.scaffold_12916 2819059 119 + 16180835 CACACACCCCGA-AAAAGGUGUGAAAUAGCCGAGGCAUGGUCUUCCAAACAAAAUAAAUAUCCAAUCGAUCAGACGAUCAAGACCGCUACGGAUAGAGCGAUUUAUGGAUAGAUAGAUAC ..((((((....-....)))))).....((....)).(((....)))...........((((((...((((....)))).....((((.(.....))))).....))))))......... ( -28.50, z-score = -2.66, R) >droWil1.scaffold_181132 824189 90 - 1035393 ---ACACAUUAAUACAAUGUGUG-AAUAACCGAAACCCUGCAACCCUUUCG--------GAGCUCUCUUUCUGAUAAUCAGAAU---UGAAAGCUAGCCAGUUUA--------------- ---(((((((.....))))))).-.....((((((...........)))))--------)((((.((.(((((.....))))).---.)).))))..........--------------- ( -21.50, z-score = -3.49, R) >consensus CACACGCCCCGA_AAAAGGUGUGAAAUUGGUGGAGCCAAGUGUUCGAAUCAAAUUAA__AGGCACUCGAAGCGACGAUCA________AUCGAUCAGGCGAUUGACAGAUAGAAAGCUGC ..((((((.........))))))...((((.....))))..................................................((((((....))))))............... ( -8.43 = -8.93 + 0.50)

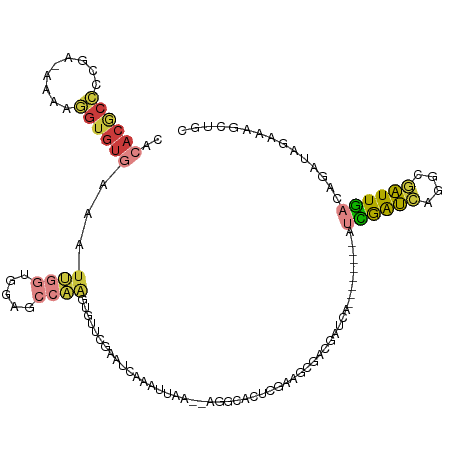
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:39 2011