
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 388,424 – 388,515 |
| Length | 91 |
| Max. P | 0.963965 |

| Location | 388,424 – 388,515 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.48028 |
| G+C content | 0.36639 |
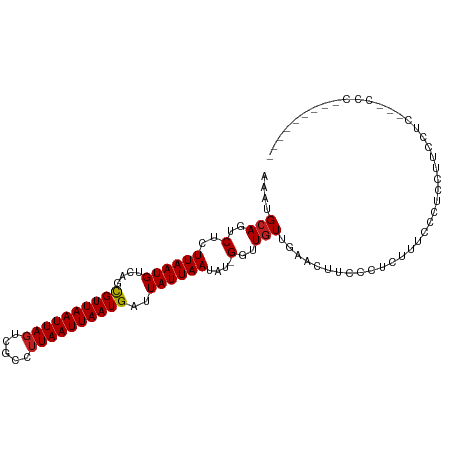
| Mean single sequence MFE | -15.76 |
| Consensus MFE | -9.64 |
| Energy contribution | -9.73 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963965 |
| Prediction | RNA |
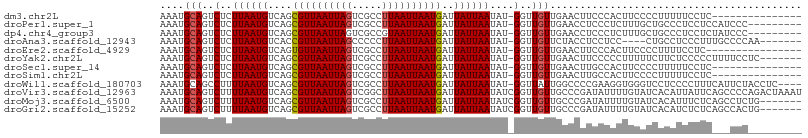
Download alignment: ClustalW | MAF
>dm3.chr2L 388424 91 - 23011544 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACUUCCCACUUCCCCUUUUUCCUC--------------- ....(((..(..((((((....((((((((((.....))))))))))..))))))...-)..)))...........................--------------- ( -12.00, z-score = -1.39, R) >droPer1.super_1 7561687 97 + 10282868 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACCUCCCUCUUUGCUGCCCUCCUCCAUCCC--------- ....(((((...((((((....((((((((((.....))))))))))..))))))...-((((....)))).........))))).............--------- ( -16.80, z-score = -2.80, R) >dp4.chr4_group3 10441929 97 + 11692001 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCGUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACCUCCCUCUUUGCUGCCCUCCUCUAUCCC--------- ....(((((...((((((....))))))((((..((((((.(((((....))))))))-)))..))))............))))).............--------- ( -17.00, z-score = -2.73, R) >droAna3.scaffold_12943 170119 95 + 5039921 AAAUGCAGUCUCUUAAUGUCACCGUUAAUUAGCCCCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUCUACCUCCUCC----CUGCCUCCCUUUGCCCCAA------- ....((((....((((((....((((((((((.....))))))))))..))))))...-(((......)))......----))))...............------- ( -13.60, z-score = -3.00, R) >droEre2.scaffold_4929 436341 90 - 26641161 AAAUGCAGUCUCUUAAUGUCAGUGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACUUCCCACUUCCCCUUUUCCUC---------------- ....(((..(..((((((....((((((((((.....))))))))))..))))))...-)..)))..........................---------------- ( -9.60, z-score = -0.45, R) >droYak2.chr2L 371984 99 - 22324452 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACUUCCCCCUUUUUCUUCUCCCCCUUUUCCUC------- ....(((..(..((((((....((((((((((.....))))))))))..))))))...-)..)))...................................------- ( -12.00, z-score = -1.75, R) >droSec1.super_14 371925 91 - 2068291 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACUUGCCACUUCCCCUUUUUCCUC--------------- ....(((..(..((((((....((((((((((.....))))))))))..))))))...-)..)))...........................--------------- ( -12.00, z-score = -0.83, R) >droSim1.chr2L 394872 91 - 22036055 AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUGUUGAACUUGCCACUUCCCCUUUUUCCUC--------------- ....(((..(..((((((....((((((((((.....))))))))))..))))))...-)..)))...........................--------------- ( -12.00, z-score = -0.83, R) >droWil1.scaffold_180703 2559641 102 + 3946847 AAAUGCAGCCUUUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU-GGUUAUUGGCCCCGAAGGUGGGUCCUCCCCUUUCAUUCUACCUC---- ......((((..((((((....((((((((((.....))))))))))..))))))...-)))).........(((((.(((....))))))))..........---- ( -23.10, z-score = -1.49, R) >droVir3.scaffold_12963 18780481 107 - 20206255 AAAUGCAGUCUUUUAAUGUCAGCGUUAAUUAGUCGGCUUAAUUAAUGAUUAUUAAUAUCGGUUGUUGCCCGAUAUUUUGUAUCACAUUAUUCAGCCCCAGACUAAAU .(((((.(.(.......).).)))))..((((((((((.(((...((((....((((((((.......))))))))....))))....))).))))...)))))).. ( -26.20, z-score = -3.52, R) >droMoj3.scaffold_6500 7968386 100 + 32352404 AAAUGCAGUCUUUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAUCGGUUGUUGCCCGAUAUUUUGUAUCACAUUUCUCAGCCUCUG------- ....((.(.((.((((((....))))))..)).)...........((((....((((((((.......))))))))....)))).........)).....------- ( -17.20, z-score = -1.53, R) >droGri2.scaffold_15252 6702245 100 - 17193109 AAAUGCAGUCUUUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAUCGGUUGUUGCCCGAUAUUUUGUAUCACAUCUCUCAGCCACUG------- .(((((.(.(.......).).)))))...((((.((.........((((....((((((((.......))))))))....)))).........)).))))------- ( -17.67, z-score = -1.46, R) >consensus AAAUGCAGUCUCUUAAUGUCAGCGUUAAUUAGUCGCCUUAAUUAAUGAUUAUUAAUAU_GGUUGUUGAACUUCCCUCUUUCCCUCCUUCCUC___CCC_________ ....(((..(..((((((....((((((((((.....))))))))))..))))))....)..))).......................................... ( -9.64 = -9.73 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:40 2011