
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,675,553 – 15,675,663 |
| Length | 110 |
| Max. P | 0.556265 |

| Location | 15,675,553 – 15,675,663 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Shannon entropy | 0.26267 |
| G+C content | 0.44540 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15675553 110 + 21146708 UCAUUAAAUUCGCCUAAGCAAA-UGCUAGCCCUGCCAU--AGAAAAGAGUAUGGGUGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((...-.))).(((...((((--(........)))))((((.(((((((((((((....)))))))))..)))).....)))))))....))))).------- ( -31.90, z-score = -1.86, R) >droGri2.scaffold_15112 2208457 101 + 5172618 UCAUUAAAUUCGCCUAAGCAGGAUGGUUGCCCUGCC------------GUGUGUGUAAGAUGAUUUAUGAAGCAGACUUCAUAAAGUUCAUAAAUGUCGCAGCAAGCAGGCGG------- ..........(((((..(((((........))))).------------.((((......(((((((((((((....)))))))))..))))......))))......))))).------- ( -30.20, z-score = -1.98, R) >droMoj3.scaffold_6496 21059153 108 + 26866924 UCAUUAAAUUCGCCUAAGCAAAAUGUUUGCCCUGCC------------GUGUGUGUAAGAUGAUUUAUGAAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGGCAAACGG .........((((((((((.....))))((..((((------------(((........(((((((((((((....)))))))))..))))......))))))).))))))))....... ( -33.34, z-score = -2.54, R) >droVir3.scaffold_12875 16621865 101 - 20611582 UCAUUAAAUUCGCCUAAGCAAAAUGUUAGCCAUGCC------------GUGUGUGUAAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((.....))).((..((((------------(((........(((((((((((((....)))))))))..))))......))))))).))))))).------- ( -31.54, z-score = -2.93, R) >droWil1.scaffold_180701 1826638 113 + 3904529 UCAUUAAAUUCGCCUAAGCAGAACGUUAUCCCUGCCCAAAUAAUGUGCGAGUGUGUGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGA------- .........((((((..((((..........))))........(((((((...((((((....(((((((((....))))))))).))))))....)))).)))...))))))------- ( -32.50, z-score = -2.10, R) >droPer1.super_2 212283 108 - 9036312 UCAUUAAAUUCGCCUAAGCAAAAUGCUAGCCCUGGCCC--CC---CGAGCAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((.....))).(((.((((..--((---(......)))...((...(((((((((....)))))))))..))......)))).)))....))))).------- ( -32.60, z-score = -1.09, R) >dp4.chr3 12800504 108 + 19779522 UCAUUAAAUUCGCCUAAGCAAAAUGCUAGCCCUGGCCC--CC---CGAGCAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((.....))).(((.((((..--((---(......)))...((...(((((((((....)))))))))..))......)))).)))....))))).------- ( -32.60, z-score = -1.09, R) >droAna3.scaffold_13266 10231719 111 - 19884421 UCAUUAAAUUCGCCUAAGCAAAAUGCUAUCCCCGCCGU--UGAAGUAAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((.....)))......(((((--.((......((((((........(((((((((....))))))))).))))))....)))))))....))))).------- ( -30.20, z-score = -1.44, R) >droEre2.scaffold_4845 9837319 110 + 22589142 UCAUUAAAUUCGCCUAAGCAAA-UGCUAGCCCUGCCAU--AGAAAAGAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((...-.))).(((((.((((--(........))))).)).((...(((((((((....)))))))))..))...........)))....))))).------- ( -31.80, z-score = -1.99, R) >droYak2.chr2R 7606842 110 - 21139217 UCAUUAAAUUCGCCUAAGCAAA-UGCUAGCCCUGCCAU--AGAAAAGAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((...-.))).(((((.((((--(........))))).)).((...(((((((((....)))))))))..))...........)))....))))).------- ( -31.80, z-score = -1.99, R) >droSec1.super_1 13165671 110 + 14215200 UCAUUAAAUUCGCCUAAGCAAA-UGCUAGCCCUGCCAU--AGAAAAGAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((...-.))).(((((.((((--(........))))).)).((...(((((((((....)))))))))..))...........)))....))))).------- ( -31.80, z-score = -1.99, R) >droSim1.chr2R 14333580 110 + 19596830 UCAUUAAAUUCGCCUAAGCAAA-UGCUAGCCCUGCCAU--AGAAAAGAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG------- ..........(((((.(((...-.))).(((((.((((--(........))))).)).((...(((((((((....)))))))))..))...........)))....))))).------- ( -31.80, z-score = -1.99, R) >consensus UCAUUAAAUUCGCCUAAGCAAAAUGCUAGCCCUGCCAU__AGAA_AGAGUAUGGGAGAGAUGAUUUAUGGAGCAGACUUCAUAAAGUUCAUAAAUGUCGCGGCAAGCAGGCGG_______ .........((((((.(((.....))).((..((((...........................(((((((((....)))))))))((.((....))..)))))).))))))))....... (-20.75 = -20.95 + 0.20)

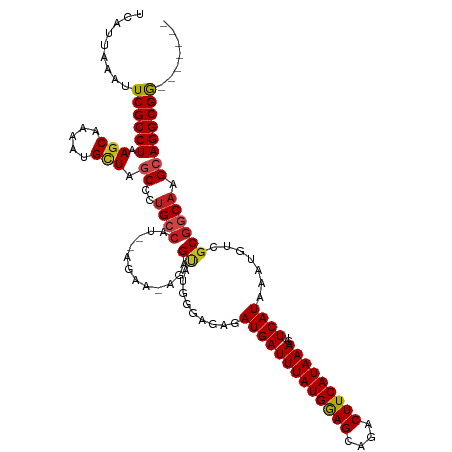
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:13 2011