| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,672,939 – 15,673,044 |
| Length | 105 |
| Max. P | 0.991652 |

| Location | 15,672,939 – 15,673,044 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.79 |
| Shannon entropy | 0.64210 |
| G+C content | 0.57097 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -12.54 |
| Energy contribution | -15.16 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
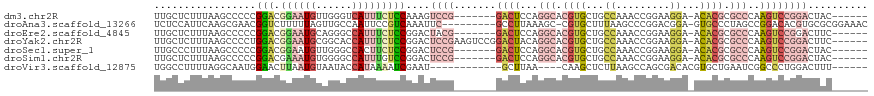
>dm3.chr2R 15672939 105 + 21146708 UUGCUCUUUAAGCCCCCGGACGGAAUGUUGGGUCAUUUCUCCAAAGUCCG-------GACUCCAGGCACGUGCUGCCAAACCGGAAGGA-ACACGCGCCCAAGUCCGGACUAC------ ..(((.....)))....(((.((((((......)))))))))..((((((-------((((...(((.((((........((....)).-.)))).)))..))))))))))..------ ( -41.00, z-score = -2.62, R) >droAna3.scaffold_13266 10229111 108 - 19884421 UCUCCAUUCAAGCGAACGGUCUUUUUAGUUGCCAAUUCCGUCAAAUUC---------GCCUUAAAGC-CGUGCUUUAAGCCCGGACGGA-GUGCCCUAGCCGGACACGUGCGCGGAAAC ......(((..(((.(((((((..((((..((..((((((((......---------((.(((((((-...)))))))))...))))))-)))).))))..)))).))).))).))).. ( -34.50, z-score = -1.57, R) >droEre2.scaffold_4845 9834635 105 + 22589142 UUGCUCUUUAAGCCCCCGGACGGAAUGCAGGGCCAUUUCUCCGGACUACG-------GACUCCAGGCACGUGCUGCCAAACCGGAAGGA-ACACGCGCCCAAGUCCGGACUUC------ ..(((.....)))..((((((((...(((.((((.....((((.....))-------)).....))).).)))..))...((....)).-............)))))).....------ ( -36.90, z-score = -1.05, R) >droYak2.chr2R 7604209 112 - 21139217 UUGCUCUUUAAGCCCCUGGACGGAAUGCGGCACCAUUUCUCCGGACUCCGAAGUCCGGACUACAGGCACGUGCUGCCAAACCGGAAGGA-ACACGCGCCCAAGUCCGGACUUC------ ..(((.....)))..(((((.((((((......))))))))))).....((((((((((((...(((.((((........((....)).-.)))).)))..))))))))))))------ ( -46.20, z-score = -3.54, R) >droSec1.super_1 13163107 105 + 14215200 UUGCCCUUUAAGCCCCCGGACGGAAUGUUGGGCCACUUCUCCGGACUCCG-------GACUCCAGGCACGUGCUGCCAAACCGGAAGGA-ACACGCGCCCAAGUCCGGACUAC------ ...............(((((.(((.((......)).))))))))..((((-------((((...(((.((((........((....)).-.)))).)))..))))))))....------ ( -36.30, z-score = -0.57, R) >droSim1.chr2R 14330174 105 + 19596830 UUGCUCUUUAAGCCCCCGGACGAAAUGUGGGGCCAUUUGUCCGGACUCCG-------GACUCCAGGCACGUGCUGCCAAACCGGAAGGA-ACACGCGCCCAAGUCCGGACUAC------ ..(((.....)))..((((((((...(((....)))))))))))..((((-------((((...(((.((((........((....)).-.)))).)))..))))))))....------ ( -41.60, z-score = -2.12, R) >droVir3.scaffold_12875 16618153 97 - 20611582 UGGCCUUUUAGGCAAUGGAACUUAAUGUAAUACCAUAAAAUCGAAU------------GCUUAA----CAAGCUCUUAAGCCAGCGACACGUGCUGAAUCGGCCCUGGACUUU------ .((((..((.((((((((.((.....))....))))....(((...------------((((((----.......))))))...)))....)))).))..)))).........------ ( -21.20, z-score = -0.41, R) >consensus UUGCUCUUUAAGCCCCCGGACGGAAUGUUGGGCCAUUUCUCCGGACUCCG_______GACUCCAGGCACGUGCUGCCAAACCGGAAGGA_ACACGCGCCCAAGUCCGGACUAC______ ...............((((((((((((......)))))).........................(((.((((........((....))...)))).)))...))))))........... (-12.54 = -15.16 + 2.62)
| Location | 15,672,939 – 15,673,044 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.79 |
| Shannon entropy | 0.64210 |
| G+C content | 0.57097 |
| Mean single sequence MFE | -45.09 |
| Consensus MFE | -16.92 |
| Energy contribution | -19.83 |
| Covariance contribution | 2.91 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
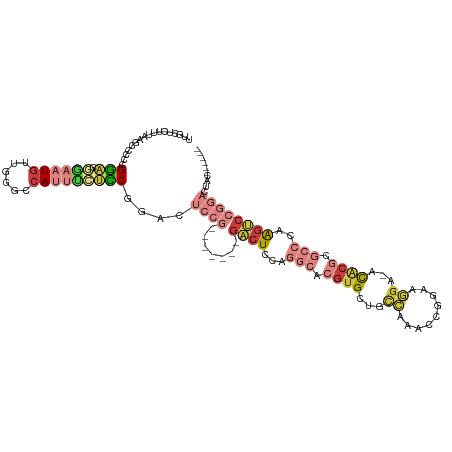
>dm3.chr2R 15672939 105 - 21146708 ------GUAGUCCGGACUUGGGCGCGUGU-UCCUUCCGGUUUGGCAGCACGUGCCUGGAGUC-------CGGACUUUGGAGAAAUGACCCAACAUUCCGUCCGGGGGCUUAAAGAGCAA ------..((((((((((..(((((((((-(((.........)).))))))))))..).)))-------))))))((((((.((((......)))).).)))))..((((...)))).. ( -47.20, z-score = -2.87, R) >droAna3.scaffold_13266 10229111 108 + 19884421 GUUUCCGCGCACGUGUCCGGCUAGGGCAC-UCCGUCCGGGCUUAAAGCACG-GCUUUAAGGC---------GAAUUUGACGGAAUUGGCAACUAAAAAGACCGUUCGCUUGAAUGGAGA (((.((......((((((.....))))))-((((((..(.((((((((...-)))))))).)---------......))))))...)).)))........((((((....))))))... ( -36.60, z-score = -0.79, R) >droEre2.scaffold_4845 9834635 105 - 22589142 ------GAAGUCCGGACUUGGGCGCGUGU-UCCUUCCGGUUUGGCAGCACGUGCCUGGAGUC-------CGUAGUCCGGAGAAAUGGCCCUGCAUUCCGUCCGGGGGCUUAAAGAGCAA ------.(((((((((((..(((((((((-(((.........)).))))))))))..).)))-------).....((((((.((((......)))).).)))))))))))......... ( -47.70, z-score = -2.38, R) >droYak2.chr2R 7604209 112 + 21139217 ------GAAGUCCGGACUUGGGCGCGUGU-UCCUUCCGGUUUGGCAGCACGUGCCUGUAGUCCGGACUUCGGAGUCCGGAGAAAUGGUGCCGCAUUCCGUCCAGGGGCUUAAAGAGCAA ------(((((((((((((((((((((((-(((.........)).)))))))))))).))))))))))))(((((.(((..........))).)))))(((....)))........... ( -56.50, z-score = -4.35, R) >droSec1.super_1 13163107 105 - 14215200 ------GUAGUCCGGACUUGGGCGCGUGU-UCCUUCCGGUUUGGCAGCACGUGCCUGGAGUC-------CGGAGUCCGGAGAAGUGGCCCAACAUUCCGUCCGGGGGCUUAAAGGGCAA ------....((((((((..(((((((((-(((.........)).))))))))))..).)))-------)))).(((((((.((((......)))).).)))))).(((.....))).. ( -50.10, z-score = -2.43, R) >droSim1.chr2R 14330174 105 - 19596830 ------GUAGUCCGGACUUGGGCGCGUGU-UCCUUCCGGUUUGGCAGCACGUGCCUGGAGUC-------CGGAGUCCGGACAAAUGGCCCCACAUUUCGUCCGGGGGCUUAAAGAGCAA ------....((((((((..(((((((((-(((.........)).))))))))))..).)))-------)))).((((((((((((......))))).))))))).((((...)))).. ( -54.40, z-score = -4.26, R) >droVir3.scaffold_12875 16618153 97 + 20611582 ------AAAGUCCAGGGCCGAUUCAGCACGUGUCGCUGGCUUAAGAGCUUG----UUAAGC------------AUUCGAUUUUAUGGUAUUACAUUAAGUUCCAUUGCCUAAAAGGCCA ------.(((.(((((((((........)).))).)))))))..(.((((.----(((.((------------(...(((((.(((......))).)))))....))).))).))))). ( -23.10, z-score = -0.31, R) >consensus ______GAAGUCCGGACUUGGGCGCGUGU_UCCUUCCGGUUUGGCAGCACGUGCCUGGAGUC_______CGGAGUCCGGAGAAAUGGCCCAACAUUCCGUCCGGGGGCUUAAAGAGCAA ........(((((.(((((((((((((((...((........))..))))))))))).)))).............(((((..((((......))))...)))))))))).......... (-16.92 = -19.83 + 2.91)

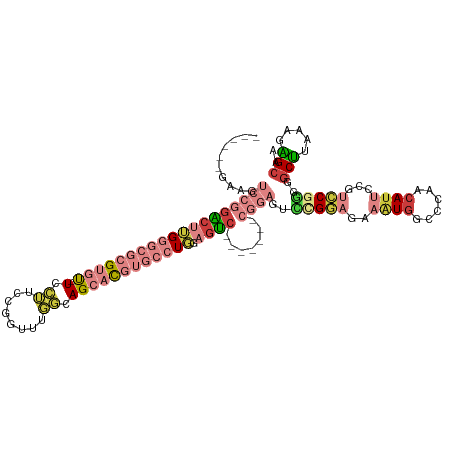
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:12 2011