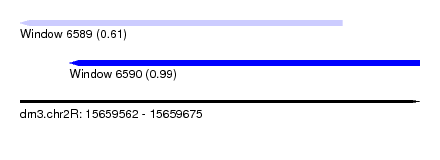
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,659,562 – 15,659,675 |
| Length | 113 |
| Max. P | 0.994907 |

| Location | 15,659,562 – 15,659,653 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.46 |
| Shannon entropy | 0.30599 |
| G+C content | 0.34777 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605633 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
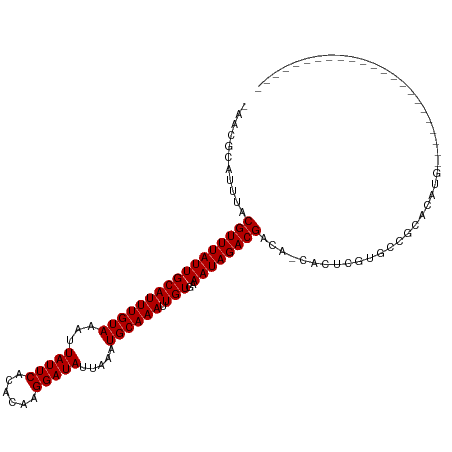
>dm3.chr2R 15659562 91 - 21146708 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGCACAUG----------------------- -...((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....((((..-...))))...)).....----------------------- ( -18.40, z-score = -1.23, R) >droSim1.chr2R 14320266 92 - 19596830 AAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGCACAUG----------------------- ....((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....((((..-...))))...)).....----------------------- ( -18.40, z-score = -1.22, R) >droSec1.super_1 13153117 92 - 14215200 AAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGCACAUG----------------------- ....((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....((((..-...))))...)).....----------------------- ( -18.40, z-score = -1.22, R) >droYak2.chr2R 7594000 91 + 21139217 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGCACAUG----------------------- -...((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....((((..-...))))...)).....----------------------- ( -18.40, z-score = -1.23, R) >droEre2.scaffold_4845 9823469 91 - 22589142 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGCACAUG----------------------- -...((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....((((..-...))))...)).....----------------------- ( -18.40, z-score = -1.23, R) >droAna3.scaffold_13266 10219096 96 + 19884421 -GACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACUCGUGCCGUACCAGAGCAC------------------ -...((.((((((....(((((((((.....(((((......))))).)))))))))..))))))....(((.((-(....))).)))......))..------------------ ( -21.10, z-score = -1.90, R) >droWil1.scaffold_180701 1799029 113 - 3904529 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CACAGACACACAACCAGU-ACGAGACUUGAGCAACAAGAA -...((.....(((((((((((((((((...(((((......))))).....)))))).)))..))))))))...-.............(.(((-.....))).).))........ ( -16.70, z-score = -0.76, R) >droVir3.scaffold_12875 16601565 84 + 20611582 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CUCGGCUACG------------------------------ -...((.((((((....(((((((((.....(((((......))))).)))))))))..)))))).....((...-..))))....------------------------------ ( -16.10, z-score = -1.81, R) >droMoj3.scaffold_6496 21038640 96 - 26866924 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA-CAUACAAGCGCUCCCUCUCACU------------------ -..(((...........(((((((((.....(((((......))))).)))))))))..((((..........))-)).....)))............------------------ ( -15.60, z-score = -1.14, R) >droGri2.scaffold_15112 2191665 97 - 5172618 -AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACGCCACGUAUACGUAUUCGUAUUCG------------------ -..........(((((((((((((((((...(((((......))))).....)))))).)))..))))))))......((.(((((....))))).))------------------ ( -18.90, z-score = -0.74, R) >consensus _AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACA_CACUCGUGCCGCACAUG_______________________ ...........(((((((((((((((((...(((((......))))).....)))))).)))..))))))))............................................ (-14.20 = -14.20 + 0.00)

| Location | 15,659,576 – 15,659,675 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Shannon entropy | 0.37425 |
| G+C content | 0.35642 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.69 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
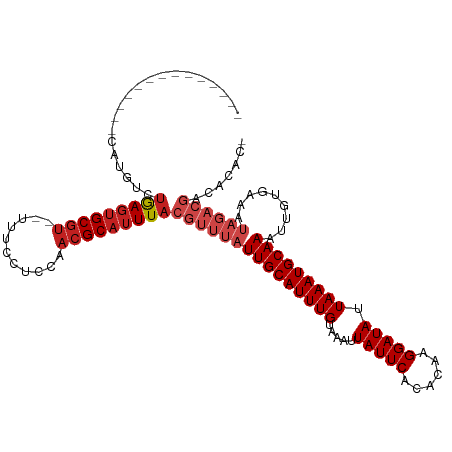
>dm3.chr2R 15659576 99 - 21146708 --------------CAUGUCUGAGUGCGU---UUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((---(......)))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.20, z-score = -3.22, R) >droSim1.chr2R 14320280 100 - 19596830 --------------CAUGUCUGAGUGCGU--UUCCUCCAAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((--((.....))))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.90, z-score = -3.44, R) >droSec1.super_1 13153131 100 - 14215200 --------------CAUGUCUGAGUGCGU--UUCCUCCAAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((--((.....))))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.90, z-score = -3.44, R) >droYak2.chr2R 7594014 99 + 21139217 --------------CAUGUCUGAGUGCGU---UUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((---(......)))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.20, z-score = -3.22, R) >droEre2.scaffold_4845 9823483 99 - 22589142 --------------CAUGUCUGAGUGCGU---UUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((---(......)))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.20, z-score = -3.22, R) >droAna3.scaffold_13266 10219115 99 + 19884421 --------------CAUGUCUGAGUGCGU---UUCCUCCGACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC- --------------..(((((((((((((---(......)))))))))).((((((((((((((((...(((((......))))).....)))))).)))..)))))))))))...- ( -26.60, z-score = -3.11, R) >droPer1.super_2 198036 79 + 9036312 --------------CAUGUCUGAGUGCGU--UUCCUCC-AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACUAGGAUAUUAAAUGCAAA--------------------- --------------......(((((((((--(......-))))))))))......(((((((((.....(((((......))))).))))))))).--------------------- ( -20.40, z-score = -3.53, R) >droWil1.scaffold_180701 1799064 100 - 3904529 --------------CAUGUCUGAGUGCGU---UUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACACA --------------..(((((((((((((---(......)))))))))).((((((((((((((((...(((((......))))).....)))))).)))..))))))))))).... ( -26.20, z-score = -3.08, R) >droVir3.scaffold_12875 16601572 115 + 20611582 CUAUCUGAGAACC-CAAGUCUGAGUGCGUCGUUGCCUAGAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACUC- .............-.......(((((((((.(..(...(((((......))))).(((((((((.....(((((......))))).)))))))))...)..)....)))).)))))- ( -27.00, z-score = -1.93, R) >droMoj3.scaffold_6496 21038659 116 - 26866924 CUAUGUGAGAGUCAUGUGUCUGAGUGCGUCGCUGCCUGGAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAU- .............((((((((((((((((..(.....)..))))))))).((((((((((((((((...(((((......))))).....)))))).)))..))))))))))))))- ( -30.60, z-score = -1.39, R) >droGri2.scaffold_15112 2191684 112 - 5172618 -----UAUGUAUGCGAACAACAACAGCAGCAUUGCCUCGAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACGCCAC -----.((((.(((...........))))))).((.(((......((((((....(((((((((.....(((((......))))).)))))))))..)))))).....))).))... ( -20.30, z-score = -0.09, R) >dp4.chr3 12786266 79 - 19779522 --------------CAUGUCUGAGUGCGU--UUCCUCC-AACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACUAGGAUAUUAAAUGCAAA--------------------- --------------......(((((((((--(......-))))))))))......(((((((((.....(((((......))))).))))))))).--------------------- ( -20.40, z-score = -3.53, R) >consensus ______________CAUGUCUGAGUGCGU__UUUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAAAUAGACGACACAC_ ....................(((((((((...........)))))))))(((((((((((((((.....(((((......))))).))))))))).........))))))....... (-17.35 = -18.69 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:10 2011