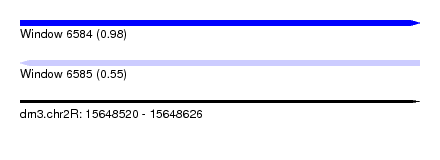
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,648,520 – 15,648,626 |
| Length | 106 |
| Max. P | 0.979581 |

| Location | 15,648,520 – 15,648,626 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.42841 |
| G+C content | 0.49744 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -23.34 |
| Energy contribution | -22.47 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
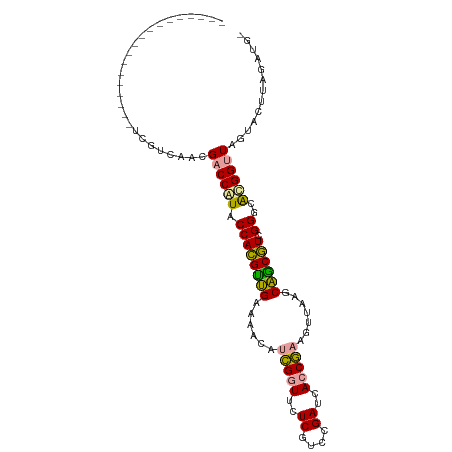
>dm3.chr2R 15648520 106 + 21146708 ACAUCUAAGUACUAACCGCGUCCGACGCUGCUUAAUUUCGGUGAUCGGACGAGAACCGAUGUAUUCAGCGUGGUAUGGUCGUUGGCGAAAGUCGAGGGGCUAAACA ........((((((..((((((((((((((........))))).))))))(((.((....)).))).)))))))))(.((.(((((....))))).)).)...... ( -35.40, z-score = -1.29, R) >anoGam1.chrU 30828652 88 - 59568033 CCACCUAAGUACUAGCCAUGCCCGAUGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACAUGGUAUGGUCGUUGACAA------------------ ........((....((((((((..((((((...(((.(((((..((....))..))))).)))..)))))))))))))).....))..------------------ ( -29.20, z-score = -2.33, R) >apiMel3.GroupUn 29499572 87 + 399230636 -CAUCCAAAUAUUAACCGUGCCCGACGCAGCUUGACUUUGCUGAUCGAACGAGAACCAAUAGUUCCUACGUGGUACGGUCGUUGAGCA------------------ -.............(((((((((((..((((........)))).))).(((.((((.....))))...))))))))))).........------------------ ( -24.10, z-score = -1.75, R) >consensus _CAUCUAAGUACUAACCGUGCCCGACGCUGCUUAACUUCGGUGAUCGGACGAGAACCGAUGUUUUCAACGUGGUAUGGUCGUUGACAA__________________ ..............((((((((..((((((...(((.(((((..((....))..))))).)))..))))))))))))))........................... (-23.34 = -22.47 + -0.87)


| Location | 15,648,520 – 15,648,626 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.42841 |
| G+C content | 0.49744 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -19.60 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15648520 106 - 21146708 UGUUUAGCCCCUCGACUUUCGCCAACGACCAUACCACGCUGAAUACAUCGGUUCUCGUCCGAUCACCGAAAUUAAGCAGCGUCGGACGCGGUUAGUACUUAGAUGU .((((((((...((....(((....))).....((((((((......(((((..((....))..))))).......)))))).)).)).)))))).))........ ( -25.52, z-score = -0.23, R) >anoGam1.chrU 30828652 88 + 59568033 ------------------UUGUCAACGACCAUACCAUGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACAUCGGGCAUGGCUAGUACUUAGGUGG ------------------......((..((((.((((((((..(((..((((..((....))..))))..)))...)))))).))..))))...)).......... ( -20.90, z-score = -0.16, R) >apiMel3.GroupUn 29499572 87 - 399230636 ------------------UGCUCAACGACCGUACCACGUAGGAACUAUUGGUUCUCGUUCGAUCAGCAAAGUCAAGCUGCGUCGGGCACGGUUAAUAUUUGGAUG- ------------------........((((((........(((((.....))))).(((((((((((........)))).)))))))))))))............- ( -25.40, z-score = -1.39, R) >consensus __________________UCGUCAACGACCAUACCACGUUGAAAACAUCGGUUCUCGUCCGAUCACCGAAGUUAAGCAGCGUCGGGCACGGUUAGUACUUAGAUG_ ..........................((((((.((((((((......(((((..((....))..))))).......)))))).))..))))))............. (-19.60 = -18.72 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:36:06 2011