| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,640,705 – 15,640,830 |
| Length | 125 |
| Max. P | 0.999027 |

| Location | 15,640,705 – 15,640,830 |
|---|---|
| Length | 125 |
| Sequences | 3 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.29250 |
| G+C content | 0.52523 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -34.70 |
| Energy contribution | -34.27 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
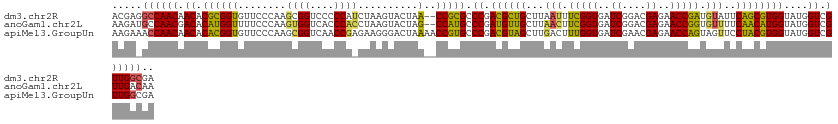
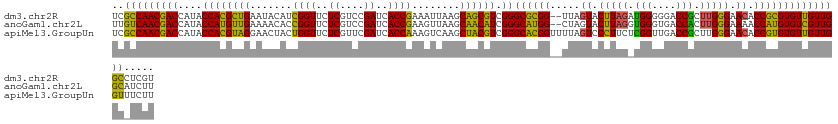
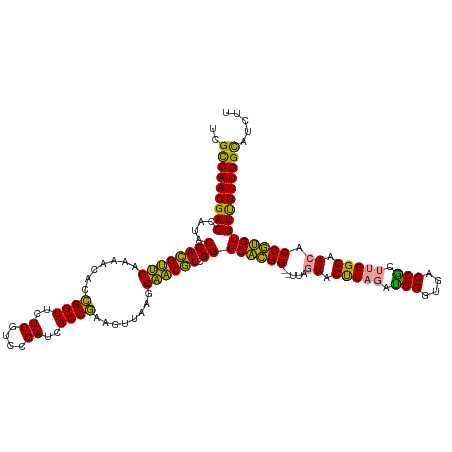
>dm3.chr2R 15640705 125 + 21146708 ACGAGGCCAACAACACGCGGUGUUCCCAAGCGGUCCCCCAUCUAAGUACUAA--CCGCGCCCGACGCUGCUUAAUUUCGGUGAUCGGACGAGAACCGAUGUAUUCAGCGUGGUAUGGUCGUUGGCGA .....((((((.((.((((((((......))((....))............)--))))).((.((((((..((...(((((..((....))..)))))..))..))))))))....)).)))))).. ( -44.30, z-score = -0.86, R) >anoGam1.chr2L 13376492 125 + 48795086 AAGAUGCCAACGACACAUGGUUUUCCCAAGUGGUCACCCACCUAAGUACUAG--CCAUGCCCGAUGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACAUGGUAUGGUCGUUGACAA ....((.(((((((.(((((((.......((((....)))).........))--))))).((.((((((...(((.(((((..((....))..))))).)))..))))))))....))))))).)). ( -41.49, z-score = -2.11, R) >apiMel3.GroupUn 283677414 127 + 399230636 AAGAAACCAACAACACACGGUGUUCCCAAGCGGUCAACCGAGAAGGGACUAAAACCGUGCCCGACGUAGCUUGACUUUGGUGAUCGAACGAGAACCAGUAGUUCCUACGUGGUAUGGUCGUUGGCGA ......(((((..........(((((....(((....)))....)))))....((((((((..((((((...(((((((((..((....))..))))).)))).)))))))))))))).)))))... ( -45.20, z-score = -2.24, R) >consensus AAGAAGCCAACAACACACGGUGUUCCCAAGCGGUCACCCAACUAAGUACUAA__CCGUGCCCGACGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACGUGGUAUGGUCGUUGGCGA .....((((((.......((....((.....))....))...............(((((((..((((((...(((.(((((..((....))..))))).)))..)))))))))))))..)))))).. (-34.70 = -34.27 + -0.43)
| Location | 15,640,705 – 15,640,830 |
|---|---|
| Length | 125 |
| Sequences | 3 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.29250 |
| G+C content | 0.52523 |
| Mean single sequence MFE | -51.51 |
| Consensus MFE | -47.41 |
| Energy contribution | -44.43 |
| Covariance contribution | -2.98 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
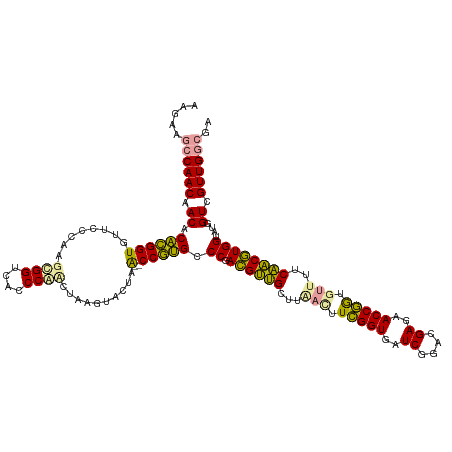
>dm3.chr2R 15640705 125 - 21146708 UCGCCAACGACCAUACCACGCUGAAUACAUCGGUUCUCGUCCGAUCACCGAAAUUAAGCAGCGUCGGGCGCGG--UUAGUACUUAGAUGGGGGACCGCUUGGGAACACCGCGUGUUGUUGGCCUCGU ..(((((((((....((((((((......(((((..((....))..))))).......)))))).))((((((--(..((.(((((.(((....))).))))).))))))))))))))))))..... ( -55.02, z-score = -3.57, R) >anoGam1.chr2L 13376492 125 - 48795086 UUGUCAACGACCAUACCAUGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACAUCGGGCAUGG--CUAGUACUUAGGUGGGUGACCACUUGGGAAAACCAUGUGUCGUUGGCAUCUU .((((((((((....((((((((..(((..((((..((....))..))))..)))...)))))).))((((((--......(((((((((....)))))))))....)))))))))))))))).... ( -52.80, z-score = -4.96, R) >apiMel3.GroupUn 283677414 127 - 399230636 UCGCCAACGACCAUACCACGUAGGAACUACUGGUUCUCGUUCGAUCACCAAAGUCAAGCUACGUCGGGCACGGUUUUAGUCCCUUCUCGGUUGACCGCUUGGGAACACCGUGUGUUGUUGGUUUCUU ..(((((((((....((((((((..(((..((((..((....))..)))).)))....)))))).))(((((((.....((((....(((....)))...))))..))))))))))))))))..... ( -46.70, z-score = -2.88, R) >consensus UCGCCAACGACCAUACCACGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACGUCGGGCACGG__UUAGUACUUAGAUGGGUGACCGCUUGGGAACACCGUGUGUUGUUGGCAUCUU ..(((((((((....((((((((.......((((..((....))..))))........)))))).))((((((.....((.(((((.(((....))).))))).)).)))))))))))))))..... (-47.41 = -44.43 + -2.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:57 2011