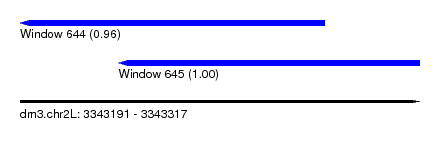
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,343,191 – 3,343,317 |
| Length | 126 |
| Max. P | 0.997863 |

| Location | 3,343,191 – 3,343,287 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 60.45 |
| Shannon entropy | 0.71672 |
| G+C content | 0.55410 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -6.97 |
| Energy contribution | -7.85 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
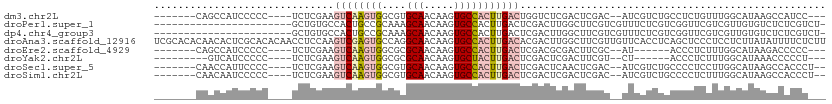
>dm3.chr2L 3343191 96 - 23011544 -------CAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUGGUCUCGACUCGAC--AUCGUCUGCCUCUGUUUGGCAUAAGCCAUCC--- -------..((((..(...----......((((((((((((.........))))))))))))(((...(((.....--...))).)))...)..))))...........--- ( -29.80, z-score = -2.12, R) >droPer1.super_1 2549693 88 + 10282868 -----------------------GCUGUGCCACUGCCGCAAAGCAACAAGUGCCACUUGACUCGACUUGGCUUCGUCGUUUCUCGUCGGUUCGUCGUUGUGUCUCUCGUCU- -----------------------...(((.((((...((...))....)))).)))..(((.((((..(((.(((.((.....)).)))...))))))).)))........- ( -19.30, z-score = 0.94, R) >dp4.chr4_group3 5452047 88 + 11692001 -----------------------GCUGUGCCACUGCCGCAAAGCAACAAGUGCCACUUGACUCGACUUGGCUUCGUCGUUUCUCGUCGGUUCGUCGUUGUGUCUCUCGUCU- -----------------------...(((.((((...((...))....)))).)))..(((.((((..(((.(((.((.....)).)))...))))))).)))........- ( -19.30, z-score = 0.94, R) >droAna3.scaffold_12916 2794023 112 + 16180835 UCGCACACAACACUCGCACACAACCUCCAAGUCGAGUGCCAGGCAACAAGUGCCACUUGACACGACUUGGCUUCGUUGUUCACCUCAGCUCCCUCCUCUUAUAUUUUCUCUU ..((...........))..(((((..((((((((.((....((((.....)))).....)).))))))))....)))))................................. ( -21.70, z-score = -1.24, R) >droEre2.scaffold_4929 3373357 90 - 26641161 -------CAGCCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCCACUUGACUCGACGCGACUUCGC--AU------ACCCUCUUUGGCAUAAGACCCCC--- -------..((((......----..((((.((((((((((((.......)))))))))))))))).(((....)))--..------........))))...........--- ( -37.20, z-score = -6.73, R) >droYak2.chr2L 3327709 88 - 22324452 ---------GUCAUCCCCC----UCUCGAAGUCAAGUGGCGCGCAACAAGUGCUACUUGACUCGACUCGACUUCGU--CU------ACCCUCUUUGGCAUAAACCCCCU--- ---------((((......----..((((.((((((((((((.......))))))))))))))))...(((...))--).------........))))...........--- ( -28.10, z-score = -5.00, R) >droSec1.super_5 1490425 97 - 5866729 -------CAACCAUUCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCAACUCGAC--AUCGUCUGCCCCUCCUUGGCAUAAGCCACCCU-- -------............----..((((.(((((((((((.........)))))))))))))))........(((--...)))..........((((....))))....-- ( -27.00, z-score = -3.34, R) >droSim1.chr2L 3281456 97 - 22036055 -------CAACAAUCCCCC----UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUCGAC--AUCGUCUGCCCCUCUUUGGCAUAAGCCACCCU-- -------............----..((((.(((((((((((.........)))))))))))))))...(((.....--...)))..........((((....))))....-- ( -28.50, z-score = -3.39, R) >consensus _______CAACCAUCCCCC____UCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUUCGC__AUCGUCUGCCCCUCUUUGGCAUAAGCCACCC___ ..............................((((((((....(((.....)))))))))))................................................... ( -6.97 = -7.85 + 0.88)

| Location | 3,343,222 – 3,343,317 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.12 |
| Shannon entropy | 0.55297 |
| G+C content | 0.58058 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.997863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
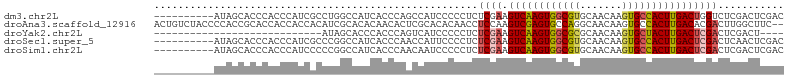
>dm3.chr2L 3343222 95 - 23011544 ----------AUAGCACCCACCCAUCGCCUGGCCAUCACCCAGCCAUCCCCCUCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUGGUCUCGACUCGAC ----------..............(((..((((.........))))........((((((((((((((((.........))))))))))))....))))..))). ( -25.50, z-score = -1.42, R) >droAna3.scaffold_12916 2794061 103 + 16180835 ACUGUCUACCCCACCGCACCACCACCACAUCGCACACAACACUCGCACACAACCUCCAAGUCGAGUGCCAGGCAACAAGUGCCACUUGACACGACUUGGCUUC-- ........................(((..(((.......((((((.((...........))))))))...((((.....))))........)))..)))....-- ( -18.40, z-score = -0.21, R) >droYak2.chr2L 3327738 73 - 22324452 ----------------------------AUAGCACCCACCCAGUCAUCCCCCUCUCGAAGUCAAGUGGCGCGCAACAAGUGCUACUUGACUCGACUCGACU---- ----------------------------..........................((((.((((((((((((.......)))))))))))))))).......---- ( -24.20, z-score = -4.12, R) >droSec1.super_5 1490457 95 - 5866729 ----------AUAGCACCCACCCAUCGCCCGGCCAUCACCCAACCAUUCCCCUCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCAACUCGAC ----------..............(((...((......))..............((((.(((((((((((.........))))))))))))))).......))). ( -22.80, z-score = -2.57, R) >droSim1.chr2L 3281488 95 - 22036055 ----------AUAGCACCCACCCAUCCCCCGGCCAUCACCCAACAAUCCCCCUCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUCGAC ----------....................((......))..............((((.(((((((((((.........)))))))))))))))........... ( -21.80, z-score = -2.28, R) >consensus __________AUAGCACCCACCCAUCGCCUGGCCAUCACCCAACCAUCCCCCUCUCGAAGUCAAGUGGCGUGCAACAAGUGCCACUUGACUCGACUCGACUCGAC ......................................................((((.((((((((((((.......))))))))))))))))........... (-17.18 = -17.78 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:37 2011