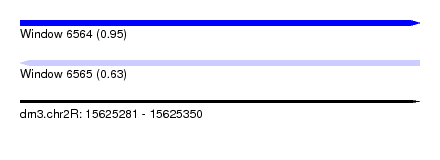
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,625,281 – 15,625,350 |
| Length | 69 |
| Max. P | 0.947391 |

| Location | 15,625,281 – 15,625,350 |
|---|---|
| Length | 69 |
| Sequences | 3 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 78.74 |
| Shannon entropy | 0.29279 |
| G+C content | 0.50725 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -18.61 |
| Energy contribution | -17.40 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
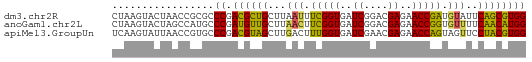
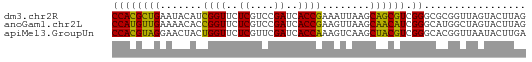
>dm3.chr2R 15625281 69 + 21146708 CUAAGUACUAACCGCGCCCGACGCUGCUUAAUUUCGGUGAUCGGACGAGAACCGAUGUAUUCAGCGUGG .................((.((((((..((...(((((..((....))..)))))..))..)))))))) ( -21.30, z-score = -1.64, R) >anoGam1.chr2L 13363180 69 + 48795086 CUAAGUACUAGCCAUGCCCGAUGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACAUGG .................((.((((((...(((.(((((..((....))..))))).)))..)))))))) ( -17.70, z-score = -0.91, R) >apiMel3.GroupUn 327590624 69 - 399230636 UCAAGUAUUAACCGUGCCCGACGUAGCUUGACUUUGGUGAUCGAACGAGAACCAGUAGUUCCUACGUGG ....((((.....))))((.((((((...(((((((((..((....))..))))).)))).)))))))) ( -18.70, z-score = -1.60, R) >consensus CUAAGUACUAACCGUGCCCGACGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACGUGG .................((.((((((...(((.(((((..((....))..))))).)))..)))))))) (-18.61 = -17.40 + -1.21)
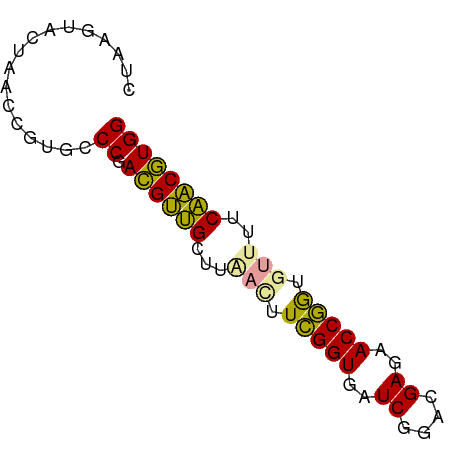
| Location | 15,625,281 – 15,625,350 |
|---|---|
| Length | 69 |
| Sequences | 3 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Shannon entropy | 0.29279 |
| G+C content | 0.50725 |
| Mean single sequence MFE | -16.31 |
| Consensus MFE | -16.10 |
| Energy contribution | -14.33 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
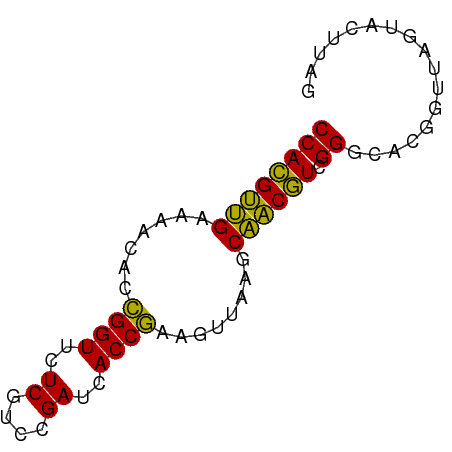
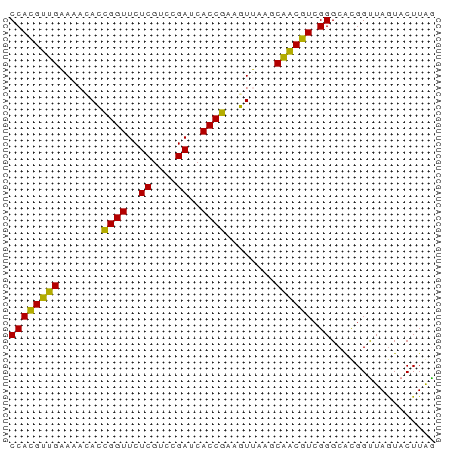
>dm3.chr2R 15625281 69 - 21146708 CCACGCUGAAUACAUCGGUUCUCGUCCGAUCACCGAAAUUAAGCAGCGUCGGGCGCGGUUAGUACUUAG ((((((((......(((((..((....))..))))).......)))))).))................. ( -18.82, z-score = -0.90, R) >anoGam1.chr2L 13363180 69 - 48795086 CCAUGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACAUCGGGCAUGGCUAGUACUUAG ((((((.........(((.......)))....((((.(((....))).))))))))))........... ( -16.20, z-score = -0.97, R) >apiMel3.GroupUn 327590624 69 + 399230636 CCACGUAGGAACUACUGGUUCUCGUUCGAUCACCAAAGUCAAGCUACGUCGGGCACGGUUAAUACUUGA ((((((((..(((..((((..((....))..)))).)))....)))))).))................. ( -13.90, z-score = 0.36, R) >consensus CCACGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACGUCGGGCACGGUUAGUACUUAG ((((((((.......((((..((....))..))))........)))))).))................. (-16.10 = -14.33 + -1.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:50 2011