| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,618,921 – 15,619,050 |
| Length | 129 |
| Max. P | 0.998388 |

| Location | 15,618,921 – 15,619,050 |
|---|---|
| Length | 129 |
| Sequences | 3 |
| Columns | 129 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Shannon entropy | 0.28279 |
| G+C content | 0.52065 |
| Mean single sequence MFE | -42.06 |
| Consensus MFE | -36.41 |
| Energy contribution | -36.20 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
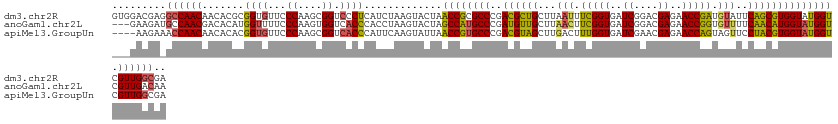
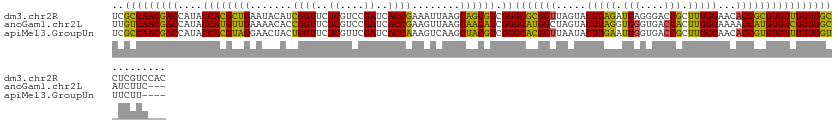
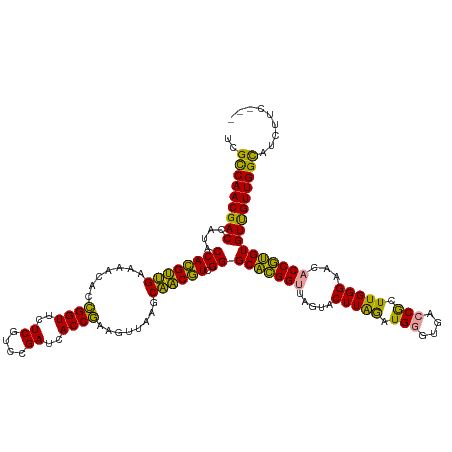
>dm3.chr2R 15618921 129 + 21146708 GUGGACGAGGCCAACAACACGCGGUGUUCCCAAGCGGUCCCUCAUCUAAGUACUAACCGCGCCCGACGCUGCUUAAUUUCGGUGAUCGGACGAGAACCGAUGUAUUCAGCGUGGUAUGGUCGUUGGCGA .........(((((((((((...))))).(((.(((((....(......).....)))))(((..((((((..((...(((((..((....))..)))))..))..))))))))).)))..)))))).. ( -43.70, z-score = -0.16, R) >anoGam1.chr2L 13355661 126 + 48795086 ---GAAGAUGCCAACGACACAUGGUUUUCCCAAGUGGUCACCCACCUAAGUACUAGCCAUGCCCGAUGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACAUGGUAUGGUCGUUGACAA ---.....((.(((((((.(((((((.......((((....)))).........))))))).((.((((((...(((.(((((..((....))..))))).)))..))))))))....))))))).)). ( -41.49, z-score = -1.98, R) >apiMel3.GroupUn 327590583 125 - 399230636 ----AAGAAACCAACAACACACGGUGUUCCCAAGCGGUCACCCAUUCAAGUAUUAACCGUGCCCGACGUAGCUUGACUUUGGUGAUCGAACGAGAACCAGUAGUUCCUACGUGGUAUGGUCGUUGGCGA ----......(((((.......((((..((.....)).)))).............((((((((..((((((...(((((((((..((....))..))))).)))).)))))))))))))).)))))... ( -41.00, z-score = -1.94, R) >consensus ___GAAGAAGCCAACAACACACGGUGUUCCCAAGCGGUCACCCAUCUAAGUACUAACCGUGCCCGACGUUGCUUAACUUCGGUGAUCGGACGAGAACCGGUGUUUUCAACGUGGUAUGGUCGUUGGCGA .........((((((.......((((...((....)).)))).............((((((((..((((((...(((.(((((..((....))..))))).)))..)))))))))))))).)))))).. (-36.41 = -36.20 + -0.21)
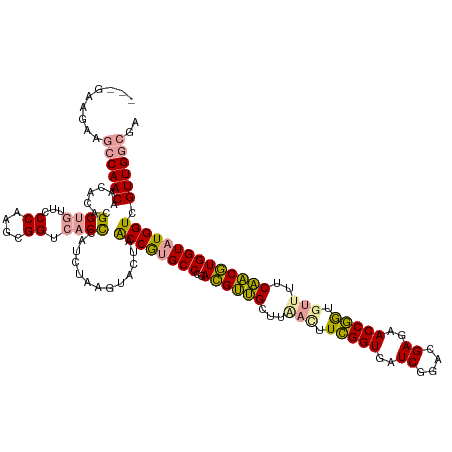
| Location | 15,618,921 – 15,619,050 |
|---|---|
| Length | 129 |
| Sequences | 3 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Shannon entropy | 0.28279 |
| G+C content | 0.52065 |
| Mean single sequence MFE | -49.11 |
| Consensus MFE | -46.21 |
| Energy contribution | -42.89 |
| Covariance contribution | -3.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15618921 129 - 21146708 UCGCCAACGACCAUACCACGCUGAAUACAUCGGUUCUCGUCCGAUCACCGAAAUUAAGCAGCGUCGGGCGCGGUUAGUACUUAGAUGAGGGACCGCUUGGGAACACCGCGUGUUGUUGGCCUCGUCCAC ..((((((((((...((((((((......(((((..((....))..))))).......)))))).))((((((((....(((....))).))))))..((.....))))).)))))))))......... ( -49.02, z-score = -2.23, R) >anoGam1.chr2L 13355661 126 - 48795086 UUGUCAACGACCAUACCAUGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACAUCGGGCAUGGCUAGUACUUAGGUGGGUGACCACUUGGGAAAACCAUGUGUCGUUGGCAUCUUC--- .((((((((((....((((((((..(((..((((..((....))..))))..)))...)))))).))((((((......(((((((((....)))))))))....)))))))))))))))).....--- ( -52.80, z-score = -4.96, R) >apiMel3.GroupUn 327590583 125 + 399230636 UCGCCAACGACCAUACCACGUAGGAACUACUGGUUCUCGUUCGAUCACCAAAGUCAAGCUACGUCGGGCACGGUUAAUACUUGAAUGGGUGACCGCUUGGGAACACCGUGUGUUGUUGGUUUCUU---- ..(((((((((....((((((((..(((..((((..((....))..)))).)))....)))))).))(((((((.....((..(.(((....))).)..))...)))))))))))))))).....---- ( -45.50, z-score = -2.56, R) >consensus UCGCCAACGACCAUACCACGUUGAAAACACCGGUUCUCGUCCGAUCACCGAAGUUAAGCAACGUCGGGCACGGUUAGUACUUAGAUGGGUGACCGCUUGGGAACACCGUGUGUUGUUGGCAUCUUC___ ..(((((((((....((((((((.......((((..((....))..))))........)))))).))(((((((.....(((((.(((....))).)))))...))))))))))))))))......... (-46.21 = -42.89 + -3.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:48 2011