| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,614,956 – 15,615,041 |
| Length | 85 |
| Max. P | 0.982947 |

| Location | 15,614,956 – 15,615,041 |
|---|---|
| Length | 85 |
| Sequences | 14 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.61097 |
| G+C content | 0.49263 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -14.06 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
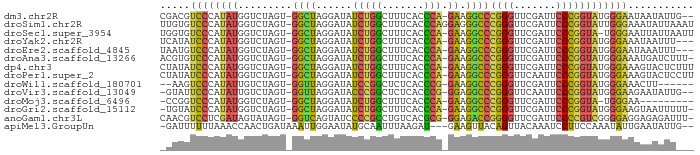
>dm3.chr2R 15614956 85 + 21146708 --CAAUAUUAUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACGUCG --.........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.60, z-score = -1.32, R) >droSim1.chr2R 14309639 88 + 19596830 AUUUAAUAUUUCCCAAUACCGGGAAUCGAACCCGGGCCCUCCUGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACACAA ..........(((((...(((((.......)))))((((....))))....((.((.....)).)).-...........)))))..... ( -23.60, z-score = -0.75, R) >droSec1.super_3954 2115 86 + 2398 AAUUAAUAAUUCCCA-UACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACACCA ..........(((((-(((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.60, z-score = -1.58, R) >droYak2.chr2R 7577931 84 - 21139217 ---AAAUUAUUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGAUAUGA ---...((((.((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....))))))).)))) ( -27.10, z-score = -1.84, R) >droEre2.scaffold_4845 9808442 84 + 22589142 ---AAAUUUAUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACAUUA ---........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.60, z-score = -1.64, R) >droAna3.scaffold_13266 9671840 86 + 19884421 -AAAGAUCAUUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACACGU -..........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.60, z-score = -1.16, R) >dp4.chr3 7002643 87 - 19779522 AAAGAGUACUUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGAUAUAG ...........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.70, z-score = -1.22, R) >droPer1.super_2 8847725 87 + 9036312 AAGGAGUACUUUCCCAUACCGGGAAUUGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGAUAUAG ...........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))..... ( -25.70, z-score = -0.75, R) >droWil1.scaffold_180701 1776931 79 + 3904529 ------AAGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-CGGGUGAGAGCCGGAUAUCCUAACC-ACUAGACAAUAUGGGACUU-- ------.....(((((((((((...((..((((((.....)-)))))..)).)))).....(((...-..)))....)))))))...-- ( -29.90, z-score = -2.31, R) >droVir3.scaffold_13049 10841829 84 - 25233164 --CAAUAUUCUUCCCAUACCGGGAAUUGAACCCGGGCCUCC-CGGGUGAGAGCCGGGUAUCCUAACC-ACUAGACAAUAUGGGAAUAC- --........((((((((((((...((..(((((((...))-)))))..)).)))).....(((...-..)))....))))))))...- ( -31.20, z-score = -2.31, R) >droMoj3.scaffold_6496 21024606 76 + 26866924 ---------UUCCCA-UACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGACCGG- ---------.(((((-(((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))....- ( -25.60, z-score = -1.10, R) >droGri2.scaffold_15112 2990100 85 - 5172618 -AAAAAUUACUUCCCAUACCGGGAAUCGAACCCGGGCCUUC-UGGGUGAAAGCCAGAUAUCCUAGCC-ACUAGACCAUAUGGGAUACA- -..........((((((((((((.......)))))....((-(((.(....))))))....((((..-.))))....)))))))....- ( -25.70, z-score = -1.78, R) >anoGam1.chr3L 36995469 86 - 41284009 -AAAUCUCUCCUCCCCGACGGGGAAUCGAACCCCGGUCUCC-CGCGUGACAGGCGGGGAUACUGACC-ACUAUACUAUCGAGGACGUUG -.......(((((......((((.......))))(((((((-(((.......)))))).....))))-...........)))))..... ( -28.90, z-score = -0.54, R) >apiMel3.GroupUn 43982512 83 + 399230636 --CAAUAUUCAAUAUUUGGAAGGAUUUGUAACUGUAACUUC---AUCUUAAAUUGCAUAUUCCAAUUUAUCAGUUGGUUUAAAAAAUC- --.(((((.((((....(((.(((.(((......))).)))---.)))...)))).)))))(((((......)))))...........- ( -8.90, z-score = 0.33, R) >consensus ___AAAUUUUUUCCCAUACCGGGAAUCGAACCCGGGCCUUC_UGGGUGAAAGCCAGAUAUCCUAGCC_ACUAGACCAUAUGGGACAUA_ ..................(((((.......)))))((((....)))).....(((......((((....))))......)))....... (-14.06 = -13.94 + -0.13)
| Location | 15,614,956 – 15,615,041 |
|---|---|
| Length | 85 |
| Sequences | 14 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.61097 |
| G+C content | 0.49263 |
| Mean single sequence MFE | -28.38 |
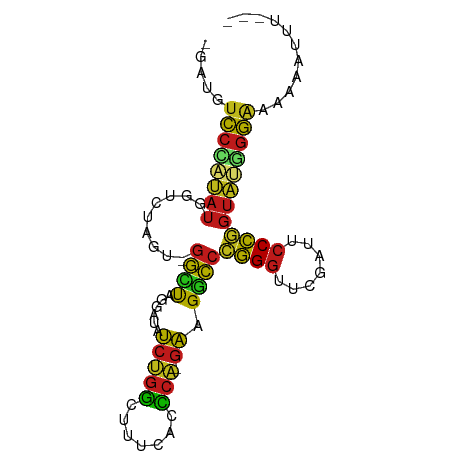
| Consensus MFE | -20.76 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
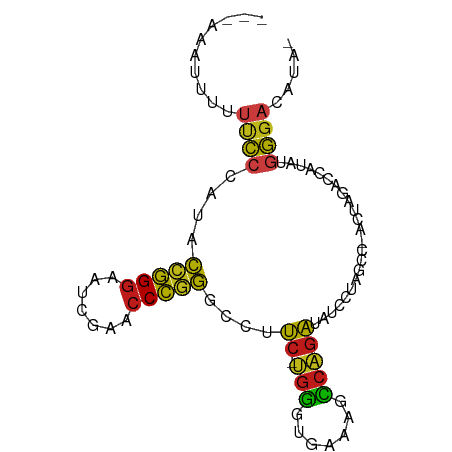
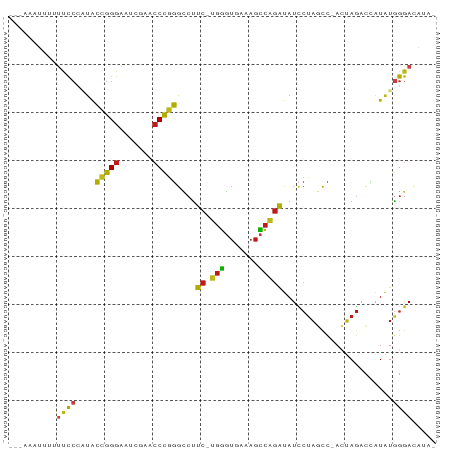
>dm3.chr2R 15614956 85 - 21146708 CGACGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAUAAUAUUG-- .....((((((((((((....-)))).........(((((((.....-)))))))((((.......)))))))))))).........-- ( -29.50, z-score = -1.29, R) >droSim1.chr2R 14309639 88 - 19596830 UUGUGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCAGGAGGGCCCGGGUUCGAUUCCCGGUAUUGGGAAAUAUUAAAU .(((.((((...(((((....-))))).((((((.(((((((......))))))).(((.......))))))))))))).)))...... ( -28.60, z-score = -0.44, R) >droSec1.super_3954 2115 86 - 2398 UGGUGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUA-UGGGAAUUAUUAAUU .....((((((((((((....-)))).........(((((((.....-)))))))((((.......)))))))-))))).......... ( -29.50, z-score = -1.05, R) >droYak2.chr2R 7577931 84 + 21139217 UCAUAUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAUAAUUU--- .....((((((((((((....-)))).........(((((((.....-)))))))((((.......))))))))))))........--- ( -29.60, z-score = -1.72, R) >droEre2.scaffold_4845 9808442 84 - 22589142 UAAUGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAUAAAUUU--- .....((((((((((((....-)))).........(((((((.....-)))))))((((.......))))))))))))........--- ( -29.50, z-score = -1.68, R) >droAna3.scaffold_13266 9671840 86 - 19884421 ACGUGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAUGAUCUUU- .(((.((((((((((((....-)))).........(((((((.....-)))))))((((.......)))))))))))).)))......- ( -31.70, z-score = -1.66, R) >dp4.chr3 7002643 87 + 19779522 CUAUAUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAGUACUCUUU .....((((((((((((....-)))).........(((((((.....-)))))))((((.......))))))))))))........... ( -29.60, z-score = -1.25, R) >droPer1.super_2 8847725 87 - 9036312 CUAUAUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAGUACUCCUU .....((((((((((((....-)))).........(((((((.....-)))))))((((.......))))))))))))........... ( -29.60, z-score = -1.30, R) >droWil1.scaffold_180701 1776931 79 - 3904529 --AAGUCCCAUAUUGUCUAGU-GGUUAGGAUAUCCGGCUCUCACCCG-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACUU------ --...(((((((.(((((...-.....))))).((((.((..(((((-(.....))))))..))...))))))))))).....------ ( -28.80, z-score = -1.19, R) >droVir3.scaffold_13049 10841829 84 + 25233164 -GUAUUCCCAUAUUGUCUAGU-GGUUAGGAUACCCGGCUCUCACCCG-GGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGAAUAUUG-- -...(((((((((((......-((....((.(((((((((((....)-))))..))))))))....)))))))))))))........-- ( -31.90, z-score = -1.67, R) >droMoj3.scaffold_6496 21024606 76 - 26866924 -CCGGUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUA-UGGGAA--------- -....((((((((((((....-)))).........(((((((.....-)))))))((((.......)))))))-))))).--------- ( -29.50, z-score = -0.97, R) >droGri2.scaffold_15112 2990100 85 + 5172618 -UGUAUCCCAUAUGGUCUAGU-GGCUAGGAUAUCUGGCUUUCACCCA-GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAGUAAUUUUU- -....((((((((((((....-)))).........(((((((.....-)))))))((((.......))))))))))))..........- ( -29.60, z-score = -1.55, R) >anoGam1.chr3L 36995469 86 + 41284009 CAACGUCCUCGAUAGUAUAGU-GGUCAGUAUCCCCGCCUGUCACGCG-GGAGACCGGGGUUCGAUUCCCCGUCGGGGAGGAGAGAUUU- ...(.((((((((........-((((......(((((.......)))-)).))))((((.......)))))))))))).)........- ( -33.80, z-score = -1.12, R) >apiMel3.GroupUn 43982512 83 - 399230636 -GAUUUUUUAAACCAACUGAUAAAUUGGAAUAUGCAAUUUAAGAU---GAAGUUACAGUUACAAAUCCUUCCAAAUAUUGAAUAUUG-- -...........((((........))))(((((.((((.......---((((...............)))).....)))).))))).-- ( -6.16, z-score = 1.07, R) >consensus _GAUGUCCCAUAUGGUCUAGU_GGCUAGGAUAUCUGGCUUUCACCCA_GAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAAAAUUU___ .....(((((((..((((.((.((((((.....))))))...))......))))(((((.......))))))))))))........... (-20.76 = -20.18 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:46 2011