| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,590,226 – 15,590,323 |
| Length | 97 |
| Max. P | 0.924535 |

| Location | 15,590,226 – 15,590,323 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.61255 |
| G+C content | 0.53965 |
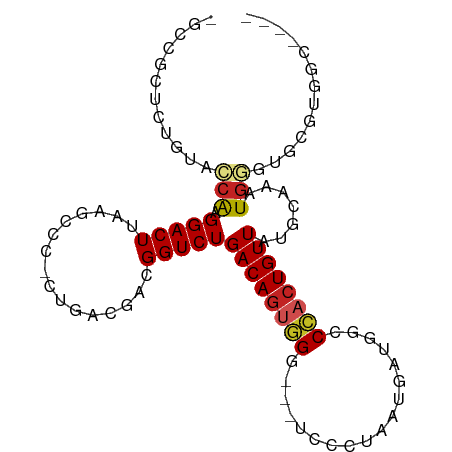
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.91 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15590226 97 + 21146708 -GCCGCUCUGUACCAGGGACUUAAGCCC-CUGACGACGGUCUGACAGUGGUUGGUCCUUAAUGACGGCCCACUGUUAUGCAAAUGGGUACGCGGCCAAA -(((((...(((((((((........))-)).......((.(((((((((.(.(((......))).).))))))))).)).....)))))))))).... ( -40.00, z-score = -2.69, R) >droSim1.chr2R 14291750 97 + 19596830 -GCCGCUCUGUACCAGGGACUUAAGCCC-CUGACGACGGUCUGACAGUGGUUGGUCCUUAAUGACGGCCCACUGUUAUGCAAAUGGGUGCGUGGCCAAA -(((((...(((((((((........))-)).......((.(((((((((.(.(((......))).).))))))))).)).....)))))))))).... ( -38.10, z-score = -1.94, R) >droSec1.super_1 13104677 97 + 14215200 -GCCGCUCUGUACCAGGGACUUAAGCCC-CUGACGACGGUCUGACAGUGGUUGGUCCUUAAUGACGGCCCACUGUUAUGCAAAUGGGUGCGUGGCCAAA -(((((...(((((((((........))-)).......((.(((((((((.(.(((......))).).))))))))).)).....)))))))))).... ( -38.10, z-score = -1.94, R) >droYak2.chr2R 7557126 97 - 21139217 -GCCGCUCUGUACCAAGGACUUAAGUCA-CUGACGACGGUCUGACAGUGGAUGGUCCUUAAUGACGGCCCACUGUUAUGCAAAUGGGUGCGUGGCCAAA -(((((...(((((..(((((...(((.-..)))...)))))((((((((.(.(((......))).).)))))))).........)))))))))).... ( -36.50, z-score = -2.36, R) >droEre2.scaffold_4845 9790500 98 + 22589142 -GCCGCUCUGUACCAAGGACUUAAGCCCUCUGACGACGGUCUGACAGUGGAAGGUCCUUAAUGACGGCCCACUGUUAUGCAAAUGGGUGCGUGGCCAAA -(((((...(((((((((((((..((..((.(((....))).))..))...)))))))).(((((((....))))))).......)))))))))).... ( -34.40, z-score = -1.52, R) >droAna3.scaffold_13266 10193067 83 - 19884421 -GGUCCUCCG-GCCGCGGACUUAAGCCCCCUGACGACGGUCUGACAGUGGG----CUCUAAUGAUGGCCCACUGUUAUGCAAAUGUACC---------- -.(((.((.(-(..((........))..)).)).))).((.((((((((((----((........)))))))))))).)).........---------- ( -31.20, z-score = -2.33, R) >dp4.chr3 12747691 85 + 19779522 -GCCACCCCG-CCCAAGGACUUAAGCCCCCUGACGACGGUCUGACAGUGGG----CUCUAAUGAUGGCCCACUGUUAUGCAAAUGGCUCAU-------- -((((..(((-(...(((..........)))...).)))..((((((((((----((........))))))))))))......))))....-------- ( -30.40, z-score = -2.56, R) >droPer1.super_2 171775 85 - 9036312 -GCCACCCCG-CCCAAGGACUUAAGCCCCCUGACGACGGUCUGACAGUGGG----CUCUAAUGAUGGCCCACUGUUAUGCAAAUGGCUCAU-------- -((((..(((-(...(((..........)))...).)))..((((((((((----((........))))))))))))......))))....-------- ( -30.40, z-score = -2.56, R) >droVir3.scaffold_12875 16564440 78 - 20611582 ------------CCAAGGACUUAA--CGUCUGAUGGCGGUCUGACAGCUGC---CAUCUAAUGAUGCCCAACUGUUAUGCAAAUGUUCAGCCAAA---- ------------....((.((.((--(((..(((((((((......)))))---))))..(((((........)))))....))))).))))...---- ( -21.10, z-score = -1.42, R) >droMoj3.scaffold_6496 20999275 79 + 26866924 -----------CGCAGGGACUUAA--CGUCUGAUGCGGGUCUGACAAUUGG---CAUCUAAUGAUCCCCAACUGUUAUGCAAAUGUUCUUUAAAA---- -----------((((.((((....--.))))..)))).((.(((((.((((---.(((....)))..)))).))))).))...............---- ( -19.70, z-score = -1.50, R) >droGri2.scaffold_15112 2161446 90 + 5172618 GCCUCUGAGAUCACAAGGACUUAA--CGCCUGAUGACGGUCUGACAGCCGC---CACGUAAUGAUGGCCAACUGUUAUGCAAAUUCUCAGCCAAA---- ....(((((((((..(((......--..)))..)))..((.((((((..((---((........))))...)))))).))....)))))).....---- ( -23.90, z-score = -1.44, R) >consensus _GCCGCUCUGUACCAAGGACUUAAGCCC_CUGACGACGGUCUGACAGUGGG___UCCCUAAUGAUGGCCCACUGUUAUGCAAAUGGGUGCGUGGC____ ............(((.(((((................)))))((((((((..................)))))))).......)))............. (-11.65 = -11.91 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:42 2011