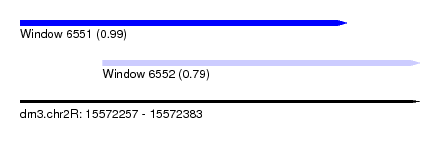
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,572,257 – 15,572,383 |
| Length | 126 |
| Max. P | 0.990792 |

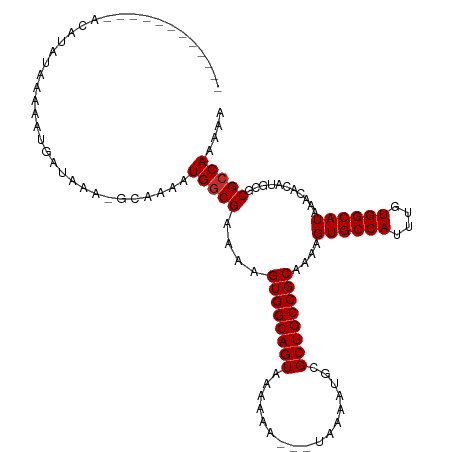
| Location | 15,572,257 – 15,572,360 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.87 |
| Shannon entropy | 0.20620 |
| G+C content | 0.37336 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -27.87 |
| Energy contribution | -27.99 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990792 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 15572257 103 + 21146708 ------------AUAUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA ------------..................-......(((((....((((((((......---........))))))))....((((((....))))))...........))))).... ( -28.94, z-score = -2.57, R) >droSec1.super_1 13086665 116 + 14215200 AUAUAUACACUAACGUAUAAAAAUGACAAAAGCAAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA ...(((((......)))))..................(((((....((((((((......---........))))))))....((((((....))))))...........))))).... ( -31.54, z-score = -2.75, R) >droYak2.chr2R 7538763 113 - 21139217 --AUAUACACUAACGUAUAGAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA --.(((((......)))))...........-......(((((....((((((((......---........))))))))....((((((....))))))...........))))).... ( -31.54, z-score = -2.70, R) >droEre2.scaffold_4845 9772665 113 + 22589142 --AUAUACACUAACGUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA --.(((((......)))))...........-......(((((....((((((((......---........))))))))....((((((....))))))...........))))).... ( -31.54, z-score = -2.76, R) >droAna3.scaffold_13266 10175746 114 - 19884421 -AUAUAUACGAUACAUAUAAAAAUGAUAAA-GCCAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA -.............................-(((((((((((....((((((((......---........)))))))).....)))))))))..((((......))))))........ ( -29.34, z-score = -2.08, R) >droWil1.scaffold_180701 1695380 109 + 3904529 ------AUAUAUACAUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA ------........................-......(((((....((((((((......---........))))))))....((((((....))))))...........))))).... ( -28.94, z-score = -2.16, R) >droVir3.scaffold_12875 16532241 105 - 20611582 ------------ACAUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAAAAAUAAUAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAA- ------------..................-......(((((....((((((((.................))))))))....((((((....))))))...........)))))...- ( -28.73, z-score = -2.31, R) >droMoj3.scaffold_6496 20969844 102 + 26866924 ------------GCAUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAAA--UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGGCAAA-- ------------((((..............-...((((((((....((((((((.......--........)))))))).....))))))))(((.......)))))))........-- ( -28.46, z-score = -1.79, R) >droGri2.scaffold_15112 2134819 103 + 5172618 ------------ACAUAUAAAAAUGAUAAA-GCAAAAUGGCGAAAAGUGGCAGUAAAAAAA--UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAA- ------------..................-......(((((....((((((((.......--........))))))))....((((((....))))))...........)))))...- ( -28.86, z-score = -2.42, R) >consensus ____________ACAUAUAAAAAUGAUAAA_GCAAAAUGGCGAAAAGUGGCAGUAAAAAA___UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAA .....................................(((((....((((((((.................))))))))....((((((....))))))...........))))).... (-27.87 = -27.99 + 0.11)

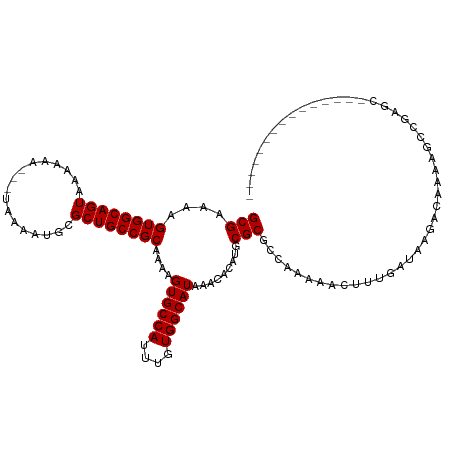
| Location | 15,572,283 – 15,572,383 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.02 |
| Shannon entropy | 0.19963 |
| G+C content | 0.43277 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.789962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15572283 100 + 21146708 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC--------------- (((....((((((((......---........))))))))....((((((....)))))).........)))...............................--------------- ( -24.64, z-score = -0.54, R) >droSec1.super_1 13086704 100 + 14215200 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC--------------- (((....((((((((......---........))))))))....((((((....)))))).........)))...............................--------------- ( -24.64, z-score = -0.54, R) >droYak2.chr2R 7538799 100 - 21139217 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC--------------- (((....((((((((......---........))))))))....((((((....)))))).........)))...............................--------------- ( -24.64, z-score = -0.54, R) >droEre2.scaffold_4845 9772701 100 + 22589142 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC--------------- (((....((((((((......---........))))))))....((((((....)))))).........)))...............................--------------- ( -24.64, z-score = -0.54, R) >droAna3.scaffold_13266 10175783 100 - 19884421 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC--------------- (((....((((((((......---........))))))))....((((((....)))))).........)))...............................--------------- ( -24.64, z-score = -0.54, R) >dp4.chr3 12701965 101 + 19779522 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAGAAAGUUUGAUAAGACAAAGGCAGUGGC-------------- (((....((((((((......---........))))))))....((((((....)))))).........)))(((.....((((....))))...)))......-------------- ( -29.94, z-score = -1.51, R) >droPer1.super_2 153578 95 - 9036312 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAGAAAGUUUGAUAAGACAAAGGC-------------------- (((....((((((((......---........))))))))....((((((....)))))).........)))(((.....((((....))))...)))-------------------- ( -28.24, z-score = -2.05, R) >droWil1.scaffold_180701 1695412 115 + 3904529 GCGAAAAGUGGCAGUAAAAAA---UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAGUUUGAUAACACACAUACACACAAAGACAAAAGACGAU (((....((((((((......---........))))))))....((((((....)))))).........)))........(((....)))............................ ( -24.84, z-score = -0.73, R) >droVir3.scaffold_12875 16532267 100 - 20611582 GCGAAAAGUGGCAGUAAAAAAAAAUAAUAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAG-UUUGAUAAGACAAAAGGCGA----------------- (((....((((((((.................))))))))....((((((....)))))).........)))(((....(-(((....))))....)))..----------------- ( -30.73, z-score = -2.62, R) >droMoj3.scaffold_6496 20969870 93 + 26866924 GCGAAAAGUGGCAGUAAAAAA--AUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGGCAAA-G-UUUGAUAAGACAAAAG--------------------- (((....((((((((......--.........))))))))....((((((....)))))).........)))......-(-(((....)))).....--------------------- ( -26.36, z-score = -1.89, R) >droGri2.scaffold_15112 2134845 94 + 5172618 GCGAAAAGUGGCAGUAAAAAA--AUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAG-UUUGAUAAGACAAAAG--------------------- (((....((((((((......--.........))))))))....((((((....)))))).........))).......(-(((....)))).....--------------------- ( -26.36, z-score = -2.14, R) >consensus GCGAAAAGUGGCAGUAAAAAA___UAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAAGACAAAAGCCGAGC_______________ (((....((((((((.................))))))))....((((((....)))))).........))).............................................. (-24.43 = -24.43 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:40 2011