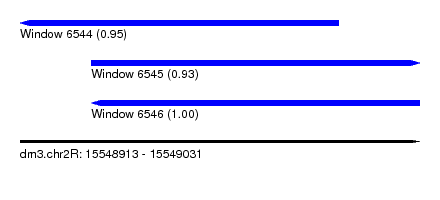
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,548,913 – 15,549,031 |
| Length | 118 |
| Max. P | 0.997460 |

| Location | 15,548,913 – 15,549,007 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Shannon entropy | 0.57644 |
| G+C content | 0.39011 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.01 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
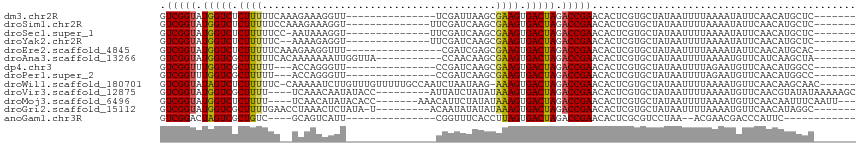
>dm3.chr2R 15548913 94 - 21146708 GUCGGUAUGGUCUCUUUUUCAAAGAAAGGUU---------------UCGAUUAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUAUUCAACAUGCUC------- .(((((.(((((((((((.....))))))((---------------(((......))))).))))).))))).....................................------- ( -17.80, z-score = -0.42, R) >droSim1.chr2R 14254347 95 - 19596830 GUCGGUAUGGUCUCUUUUUCCAAAGAAAGGU--------------UUCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUAUUCAACAUGCUC------- .(((((.(((((((((((......))))))(--------------((((......))))).))))).))))).....................................------- ( -18.00, z-score = -0.37, R) >droSec1.super_1 13063642 94 - 14215200 GUCGGUAUGGUCUCUUUUUCC-AAUAAAGGU--------------UUCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUAUUCAACAUGCUC------- .(((((.(((((.(((((...-...)))))(--------------((((......))))).))))).))))).....................................------- ( -16.50, z-score = -0.30, R) >droYak2.chr2R 7515343 93 + 21139217 GUCGGUAUGGUCUCUUUUUC--AAAAGAGGU--------------UUCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUAUUCAACAUGCUC------- .(((((.((((((((((...--.)))))..(--------------((((......))))).))))).))))).....................................------- ( -17.30, z-score = -0.20, R) >droEre2.scaffold_4845 9749165 93 - 22589142 GUCGGUAUGGUCUCUUUUUCAAAGAAGGUUU----------------CGAUCGAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUAUUCAACAUGCAC------- .(((((.(((((((((((....))))))(((----------------((......))))).))))).)))))......((((.......................))))------- ( -19.80, z-score = -0.66, R) >droAna3.scaffold_13266 10152946 98 + 19884421 GUCGGUAUGGUCGCUUUUUCACAAAAAAAUUGGUUA-----------CCAACAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAUCAAGCUA------- .(((((.((((((((((....(((.....)))(((.-----------......))))))))))))).))))).....................................------- ( -19.20, z-score = -0.79, R) >dp4.chr3 12677575 91 - 19779522 GUCGGUUUGGUCGCUUUUU---ACCAGGGUU---------------CCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAGAAUGUUCAACAUGGCC------- (((((((((((((((((..---....((...---------------))........))))))))))))))(((((.((..............)).))))).....))).------- ( -26.98, z-score = -2.08, R) >droPer1.super_2 130222 91 + 9036312 GUCGGUUUGGUCGCUUUUU---ACCAGGGUU---------------CCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAGAAUGUUCAACAUGGCC------- (((((((((((((((((..---....((...---------------))........))))))))))))))(((((.((..............)).))))).....))).------- ( -26.98, z-score = -2.08, R) >droWil1.scaffold_180701 1651735 108 - 3904529 GUCGGUAUAGUCUCUUUUUC-CAAAAAUCUUGUUUGUUUUUGCCAAUCUAAUAAG-AAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAACAAGCAAC------ .(((((.(((((.((((((.-(((((((.......)))))))...........))-)))).))))).))))).......((((.....................))))..------ ( -19.30, z-score = -1.48, R) >droVir3.scaffold_12875 16499334 103 + 20611582 GUCGGUAUGGUCGCUUUU----UCAAACAAUAUACC---------AUUAUCUAUAUAAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAACGUAUAUAAAAAGC .(((((.((((((((((.----.......(((((..---------......))))))))))))))).)))))........(((....(((((..(((.....)))...)))))))) ( -18.60, z-score = -1.64, R) >droMoj3.scaffold_6496 20936768 102 - 26866924 GUCGGUAUGGUCUCUUUU----UCAACAUAUACACC-------AAACAUUCUAUAUAAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAACAAUUUCAAUU--- .(((((.(((((.((((.----..............-------.............)))).))))).))))).........................................--- ( -14.19, z-score = -1.04, R) >droGri2.scaffold_15112 2108803 99 - 5172618 GUCGGUAUGGUCGCUUUUGAACCUAAACUCUAUA-U---------ACAAUAUAUAUAAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAACAUAGGC------- ((((((.(((((((((..((........))((((-(---------(.....))))))))))))))).)))(((((....................)))))......)))------- ( -18.95, z-score = -1.39, R) >anoGam1.chr3R 43698391 83 - 53272125 GUCGGACUAGUCGCUGUC----GCAGUCAUU---------------CGGUUUCACCUUAGUGACUAGACCGAACACUCGCGUCCUAA--ACGAACGACCCAUUC------------ (((((((........)))----(((((..((---------------(((((((((....))))...))))))).))).)).......--.....))))......------------ ( -22.40, z-score = -1.12, R) >consensus GUCGGUAUGGUCUCUUUUUC__AAAAAAGUU_______________UCGAUCAAGCGAAGUGACUAGACCGAACACUCGUGCUAUAAUUUUAAAAUGUUCAACAUGCUC_______ .(((((.(((((.((((.......................................)))).))))).)))))............................................ (-13.32 = -13.01 + -0.31)
| Location | 15,548,934 – 15,549,031 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Shannon entropy | 0.50064 |
| G+C content | 0.38832 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.59 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
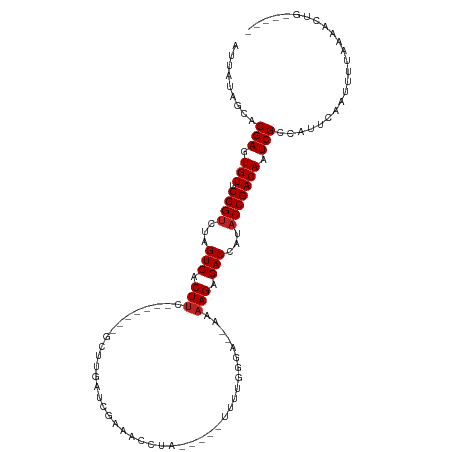
>dm3.chr2R 15548934 97 + 21146708 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUAAUCGAAACCUU-----UCUUU-GA--AAAAGAGACCAUACCGACAUUCGCCAUUCAAUUUUAAAACUG----- .........(((((((.((((...(((..(((-------(......)))).....-----((((.-..--..)))))))...)))))))))))...................----- ( -19.50, z-score = -2.22, R) >droSim1.chr2R 14254368 98 + 19596830 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGAUCGAAACCUU-----UCUUUGGA--AAAAGAGACCAUACCGACAUUCGCCAUUCAAUUUUAAAACUG----- .........(((((((.((((...(((..(((-------(......)))).....-----(((((...--.))))))))...)))))))))))...................----- ( -23.00, z-score = -2.38, R) >droSec1.super_1 13063663 97 + 14215200 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGAUCGAAACCUU-----UAUUG-GA--AAAAGAGACCAUACCGACAUUCGCCAUUCAAUUUUAAAACUG----- .........(((((((.((((...(((..(((-------(......)))).((..-----....)-).--......)))...)))))))))))...................----- ( -20.40, z-score = -1.66, R) >droYak2.chr2R 7515364 96 - 21139217 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGAUCGAAACCUC-----UUUU--GA--AAAAGAGACCAUACCGACAUUCGCCAUUCAAUUUUAAAACUG----- .........(((((((((((((.(((......-------))).))))))....((-----((((--..--..))))))........)))))))...................----- ( -20.90, z-score = -2.14, R) >droEre2.scaffold_4845 9749186 96 + 22589142 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUCGAUCGAAACCUU-----CUUU--GA--AAAAGAGACCAUACCGACAUUCGCCAUUCAAUUUUAAAACUG----- .........(((((((.((((...(((..(((-------(......))))....(-----(((.--..--..)))))))...)))))))))))...................----- ( -19.50, z-score = -1.53, R) >droAna3.scaffold_13266 10152967 101 - 19884421 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGUUGGUAACCAAUUU--UUUUGUGA--AAAAGCGACCAUACCGACAAUCGCCAUUCAAUUUUAAAAUUC----- .........(((.(((.((((...(((.((((-------((..(((((...)))))..--....))))--...)).)))...))))))).)))...................----- ( -19.70, z-score = -1.09, R) >dp4.chr3 12677596 94 + 19779522 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGAUCGGAACC---------CUGGUA--AAAAGCGACCAAACCGACAAUCGCCAUUUAAUUUUAAAAUUU----- .........(((.(((.((((.........((-------((((.(((((....---------))))).--..))))))....))))))).)))...................----- ( -19.72, z-score = -1.14, R) >droPer1.super_2 130243 94 - 9036312 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC-------GCUUGAUCGGAACC---------CUGGUA--AAAAGCGACCAAACCGACAAUCGCCAUUUAAUUUUAAAAUUU----- .........(((.(((.((((.........((-------((((.(((((....---------))))).--..))))))....))))))).)))...................----- ( -19.72, z-score = -1.14, R) >droWil1.scaffold_180701 1651757 115 + 3904529 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUUCUUAUUAGAUUGGCAAAAACAAACAAGAUUUUUGGA--AAAAGAGACUAUACCGACAAUCGCCAUUCAAUUUUAAAAGUAAUUUC .........(((.(((.((((.(((((.(..(((....)))..).(((((((......).))))))..--......))))).))))))).)))........................ ( -21.50, z-score = -1.55, R) >droVir3.scaffold_12875 16499362 99 - 20611582 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUU------AUAUAGAUAAUGGUAUAUUG---UUUGA----AAAAGCGACCAUACCGACAAUCGCCAUUUAAUUUUAAAAUAC----- .........(((.(((.((((...(((.((((------...(((((((((...)))))---)))).----.)))).)))...))))))).)))...................----- ( -19.40, z-score = -1.58, R) >droGri2.scaffold_15112 2108824 102 + 5172618 AUUAUAGCACGAGUGUUCGGUCUAGUCACUUU------AUAUAUAUUGUA-UAUAGAG---UUUAGGUUCAAAAGCGACCAUACCGACAAUCGCCAUUUAAUUUUAAAAUAC----- .........(((.(((.((((...((((((((------((((((...)))-)))))))---)....((......)))))...))))))).)))...................----- ( -20.90, z-score = -2.26, R) >anoGam1.chr3R 43698406 75 + 53272125 -UUAGGACGCGAGUGUUCGGUCUAGUCACUAA-----------GGUGAAACCGAA--------UGACUGCGACAGCGACUAGUCCGACAAUCGCC---------------------- -.......((((.(((.(((.((((((.....-----------((.....))...--------.....((....)))))))).)))))).)))).---------------------- ( -27.60, z-score = -2.67, R) >consensus AUUAUAGCACGAGUGUUCGGUCUAGUCACUUC_______GCUUGAUCGAAACCUA_____UUUUGGGA__AAAAGAGACCAUACCGACAAUCGCCAUUCAAUUUUAAAACUG_____ .........(((.(((.((((...(((.(((.........................................))).)))...))))))).)))........................ (-11.42 = -11.59 + 0.17)
| Location | 15,548,934 – 15,549,031 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Shannon entropy | 0.50064 |
| G+C content | 0.38832 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15548934 97 - 21146708 -----CAGUUUUAAAAUUGAAUGGCGAAUGUCGGUAUGGUCUCUUUU--UC-AAAGA-----AAGGUUUCGAUUAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----(((((....)))))....((((.(((((((.(((((((((((--..-...))-----))))(((((......)-------)))).))))).)))).))).))))........ ( -24.80, z-score = -1.89, R) >droSim1.chr2R 14254368 98 - 19596830 -----CAGUUUUAAAAUUGAAUGGCGAAUGUCGGUAUGGUCUCUUUU--UCCAAAGA-----AAGGUUUCGAUCAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----(((((....)))))....((((.(((((((.(((((((((((--......))-----))))(((((......)-------)))).))))).)))).))).))))........ ( -25.00, z-score = -1.65, R) >droSec1.super_1 13063663 97 - 14215200 -----CAGUUUUAAAAUUGAAUGGCGAAUGUCGGUAUGGUCUCUUUU--UC-CAAUA-----AAGGUUUCGAUCAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----(((((....)))))....((((.(((((((.(((((.(((((--..-....)-----))))(((((......)-------)))).))))).)))).))).))))........ ( -23.50, z-score = -1.42, R) >droYak2.chr2R 7515364 96 + 21139217 -----CAGUUUUAAAAUUGAAUGGCGAAUGUCGGUAUGGUCUCUUUU--UC--AAAA-----GAGGUUUCGAUCAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----(((((....)))))....((((.(((((((.((((((((((.--..--.)))-----))..(((((......)-------)))).))))).)))).))).))))........ ( -24.30, z-score = -1.50, R) >droEre2.scaffold_4845 9749186 96 - 22589142 -----CAGUUUUAAAAUUGAAUGGCGAAUGUCGGUAUGGUCUCUUUU--UC--AAAG-----AAGGUUUCGAUCGAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----(((((....)))))....((((.(((((((.(((((((((((--..--..))-----))))(((((......)-------)))).))))).)))).))).))))........ ( -24.20, z-score = -1.10, R) >droAna3.scaffold_13266 10152967 101 + 19884421 -----GAAUUUUAAAAUUGAAUGGCGAUUGUCGGUAUGGUCGCUUUU--UCACAAAA--AAAUUGGUUACCAACAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----..................((((.(((((((.((((((((((.--...(((..--...)))(((.......)))-------)))))))))).)))).))).))))........ ( -25.40, z-score = -2.25, R) >dp4.chr3 12677596 94 - 19779522 -----AAAUUUUAAAAUUAAAUGGCGAUUGUCGGUUUGGUCGCUUUU--UACCAG---------GGUUCCGAUCAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----..................((((.((((((((((((((((((.--.....(---------(...))........-------))))))))))))))).))).))))........ ( -28.94, z-score = -3.05, R) >droPer1.super_2 130243 94 + 9036312 -----AAAUUUUAAAAUUAAAUGGCGAUUGUCGGUUUGGUCGCUUUU--UACCAG---------GGUUCCGAUCAAGC-------GAAGUGACUAGACCGAACACUCGUGCUAUAAU -----..................((((.((((((((((((((((((.--.....(---------(...))........-------))))))))))))))).))).))))........ ( -28.94, z-score = -3.05, R) >droWil1.scaffold_180701 1651757 115 - 3904529 GAAAUUACUUUUAAAAUUGAAUGGCGAUUGUCGGUAUAGUCUCUUUU--UCCAAAAAUCUUGUUUGUUUUUGCCAAUCUAAUAAGAAAGUGACUAGACCGAACACUCGUGCUAUAAU .......................((((.(((((((.(((((.(((((--(.(((((((.......)))))))...........)))))).))))).)))).))).))))........ ( -24.20, z-score = -2.20, R) >droVir3.scaffold_12875 16499362 99 + 20611582 -----GUAUUUUAAAAUUAAAUGGCGAUUGUCGGUAUGGUCGCUUUU----UCAAA---CAAUAUACCAUUAUCUAUAU------AAAGUGACUAGACCGAACACUCGUGCUAUAAU -----..................((((.(((((((.((((((((((.----.....---..(((((........)))))------)))))))))).)))).))).))))........ ( -24.50, z-score = -3.37, R) >droGri2.scaffold_15112 2108824 102 - 5172618 -----GUAUUUUAAAAUUAAAUGGCGAUUGUCGGUAUGGUCGCUUUUGAACCUAAA---CUCUAUA-UACAAUAUAUAU------AAAGUGACUAGACCGAACACUCGUGCUAUAAU -----..................((((.(((((((.(((((((((..((.......---.))((((-((.....)))))------)))))))))).)))).))).))))........ ( -24.70, z-score = -3.34, R) >anoGam1.chr3R 43698406 75 - 53272125 ----------------------GGCGAUUGUCGGACUAGUCGCUGUCGCAGUCA--------UUCGGUUUCACC-----------UUAGUGACUAGACCGAACACUCGCGUCCUAA- ----------------------.((((.((((((.((((((((((.....(...--------..)((.....))-----------.)))))))))).))).))).)))).......- ( -27.70, z-score = -2.71, R) >consensus _____CAAUUUUAAAAUUGAAUGGCGAUUGUCGGUAUGGUCGCUUUU__UCCCAAAA_____UAGGUUUCGAUCAAGC_______GAAGUGACUAGACCGAACACUCGUGCUAUAAU .......................((((.(((((((.(((((.((((.......................................)))).))))).)))).))).))))........ (-19.52 = -19.16 + -0.37)
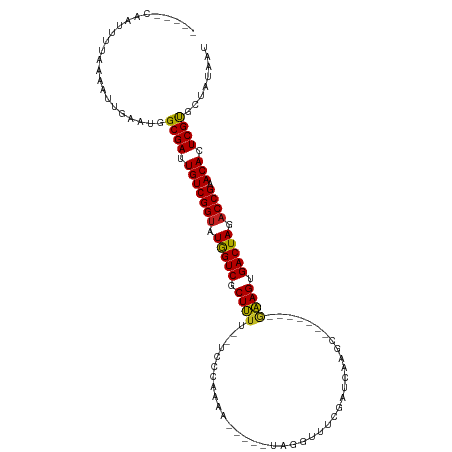
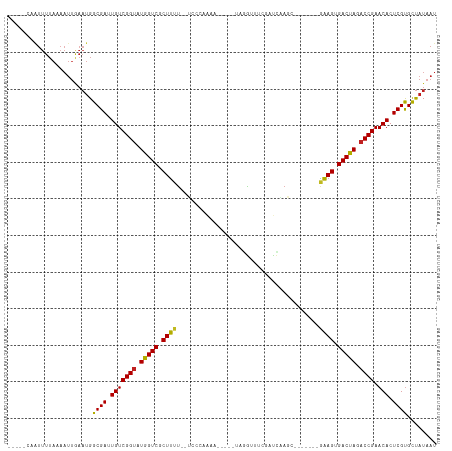
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:35 2011