| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,548,794 – 15,548,885 |
| Length | 91 |
| Max. P | 0.999328 |

| Location | 15,548,794 – 15,548,885 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.23 |
| Shannon entropy | 0.30940 |
| G+C content | 0.41513 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.09 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.12 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
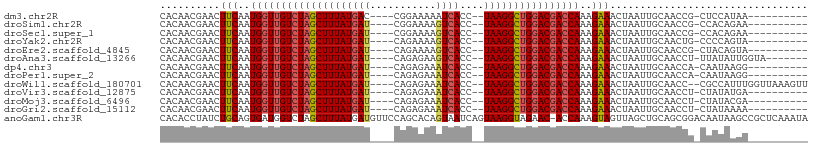
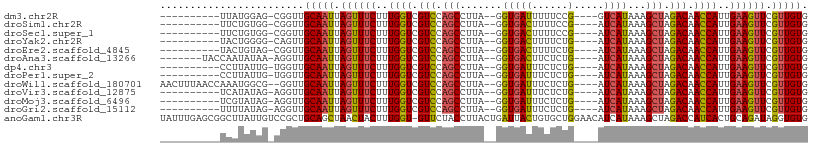
>dm3.chr2R 15548794 91 + 21146708 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAC----CGGAAAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCG-CUCCAUAA---------- ..........(((..(((((((((((((((((((.----........)))..--))))))))))))))))..))).......((....)-).......---------- ( -25.00, z-score = -4.15, R) >droSim1.chr2R 14254228 91 + 19596830 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CGGAAAAGUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCG-CCACAGAA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..))).......((....)-).......---------- ( -26.60, z-score = -3.71, R) >droSec1.super_1 13063523 91 + 14215200 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CGGAAAAGUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCG-CCACAGAA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..))).......((....)-).......---------- ( -26.60, z-score = -3.71, R) >droYak2.chr2R 7515224 91 - 21139217 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAAAAGUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACUG-CCCCAGUA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))..........((((-...)))).---------- ( -27.70, z-score = -3.98, R) >droEre2.scaffold_4845 9749045 91 + 22589142 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAAAAGUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCG-CUACAGUA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))(((....((....)-)...))).---------- ( -27.90, z-score = -4.31, R) >droAna3.scaffold_13266 10152819 94 - 19884421 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAGUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCU-UUAUAUUGGUA------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))..........(((.-.......))).------- ( -28.90, z-score = -4.12, R) >dp4.chr3 12677418 91 + 19779522 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCA-CAAUAAGG---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..))).((.((((......-)))).)).---------- ( -28.20, z-score = -5.44, R) >droPer1.super_2 130065 91 - 9036312 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCA-CAAUAAGG---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..))).((.((((......-)))).)).---------- ( -28.20, z-score = -5.44, R) >droWil1.scaffold_180701 1651610 100 + 3904529 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACC--CGCCAUUUGGUUAAAGUU .......(((((...((((((((((((((((((((----.......))))..--))))))))))))))))....((((((..((....--.))...)))))).))))) ( -31.00, z-score = -4.35, R) >droVir3.scaffold_12875 16499190 91 - 20611582 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCU-CUAUAUGA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))..............-........---------- ( -26.20, z-score = -4.44, R) >droMoj3.scaffold_6496 20936627 91 + 26866924 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCU-CUAUACGA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))..............-........---------- ( -26.20, z-score = -4.57, R) >droGri2.scaffold_15112 2108672 91 + 5172618 CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU----CAGAGAAAUCACC--UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCU-CUAUAAAA---------- ..........(((..((((((((((((((((((((----.......))))..--))))))))))))))))..)))..............-........---------- ( -26.20, z-score = -4.94, R) >anoGam1.chr3R 43698209 107 + 53272125 CACACCUAUCUGCAGUGAUGGUCUAGCUUUAUGAUGUUCCAGCACAGUAAUCAGUAAGGUAGAAC-ACCAAAGUAGUUAGCUGCAGCGGACAAUAAGCCGCUCAAAUA ..((.(((.((((..((.((.((((.(((..((((..............))))..))).)))).)-).))..)))).))).)).(((((........)))))...... ( -25.34, z-score = -0.39, R) >consensus CACAACGAACUUCAAUGGUUGUCUAGCUUUAUGAU____CAGAGAAAUCACC__UAAGGCUGGACGACCAAAGAAACUAAUUGCAACCG_CUAUAAGA__________ ..........(((..((((((((((((((((((((...........))))....))))))))))))))))..)))................................. (-23.06 = -23.09 + 0.04)
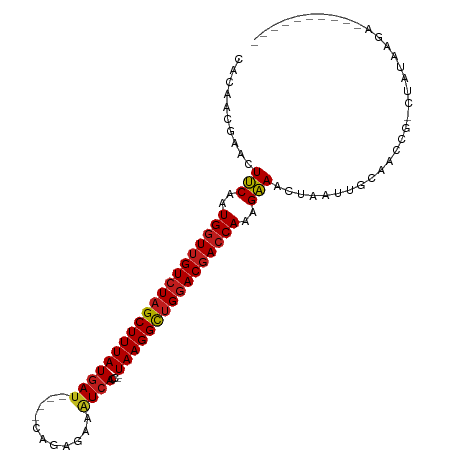
| Location | 15,548,794 – 15,548,885 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.23 |
| Shannon entropy | 0.30940 |
| G+C content | 0.41513 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15548794 91 - 21146708 ----------UUAUGGAG-CGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUUUCCG----GUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -23.80, z-score = -1.69, R) >droSim1.chr2R 14254228 91 - 19596830 ----------UUCUGUGG-CGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGACUUUUCCG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.(((((......)----.))))...))).))).))))..)))..).))))). ( -21.80, z-score = -0.86, R) >droSec1.super_1 13063523 91 - 14215200 ----------UUCUGUGG-CGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGACUUUUCCG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.(((((......)----.))))...))).))).))))..)))..).))))). ( -21.80, z-score = -0.86, R) >droYak2.chr2R 7515224 91 + 21139217 ----------UACUGGGG-CAGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGACUUUUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------.((((...-)))).(((((.(..(((..((((.(((.(((....--.(((((......)----.))))...))).))).))))..)))..).))))). ( -22.90, z-score = -0.82, R) >droEre2.scaffold_4845 9749045 91 - 22589142 ----------UACUGUAG-CGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGACUUUUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.(((((......)----.))))...))).))).))))..)))..).))))). ( -22.40, z-score = -1.22, R) >droAna3.scaffold_13266 10152819 94 + 19884421 -------UACCAAUAUAA-AGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGACUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG -------.(((.......-.))).(((((.(..(((..((((.(((.(((....--.(((((......)----.))))...))).))).))))..)))..).))))). ( -23.60, z-score = -2.15, R) >dp4.chr3 12677418 91 - 19779522 ----------CCUUAUUG-UGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -25.00, z-score = -2.93, R) >droPer1.super_2 130065 91 + 9036312 ----------CCUUAUUG-UGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -25.00, z-score = -2.93, R) >droWil1.scaffold_180701 1651610 100 - 3904529 AACUUUAACCAAAUGGCG--GGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ......((((........--))))(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -27.60, z-score = -2.26, R) >droVir3.scaffold_12875 16499190 91 + 20611582 ----------UCAUAUAG-AGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -25.00, z-score = -2.59, R) >droMoj3.scaffold_6496 20936627 91 - 26866924 ----------UCGUAUAG-AGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -25.00, z-score = -2.57, R) >droGri2.scaffold_15112 2108672 91 - 5172618 ----------UUUUAUAG-AGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA--GGUGAUUUCUCUG----AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ----------........-.....(((((.(..(((..((((.(((.(((....--.((((((.....)----)))))...))).))).))))..)))..).))))). ( -25.00, z-score = -2.77, R) >anoGam1.chr3R 43698209 107 - 53272125 UAUUUGAGCGGCUUAUUGUCCGCUGCAGCUAACUACUUUGGU-GUUCUACCUUACUGAUUACUGUGCUGGAACAUCAUAAAGCUAGACCAUCACUGCAGAUAGGUGUG ......((((((.....).)))))(((.(((.((.(..((((-(.((((.(((..(((....(((......))))))..))).)))).)))))..).)).))).))). ( -27.00, z-score = -0.39, R) >consensus __________UCAUAUAG_CGGUUGCAAUUAGUUUCUUUGGUCGUCCAGCCUUA__GGUGAUUUCUCUG____AUCAUAAAGCUAGACAACCAUUGAAGUUCGUUGUG ........................(((((.((((((..((((.(((.(((.......((((.............))))...))).))).))))..)))))).))))). (-17.94 = -18.05 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:32 2011