| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,548,488 – 15,548,591 |
| Length | 103 |
| Max. P | 0.999751 |

| Location | 15,548,488 – 15,548,591 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.64 |
| Shannon entropy | 0.60533 |
| G+C content | 0.37849 |
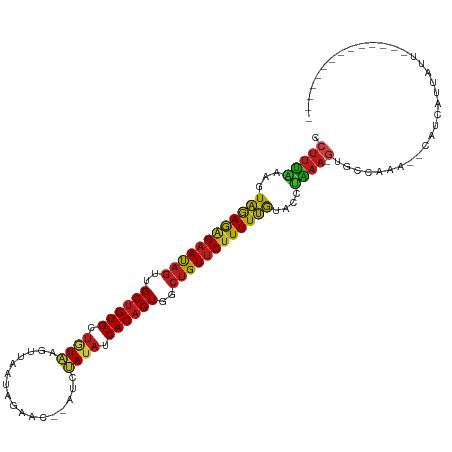
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.42 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
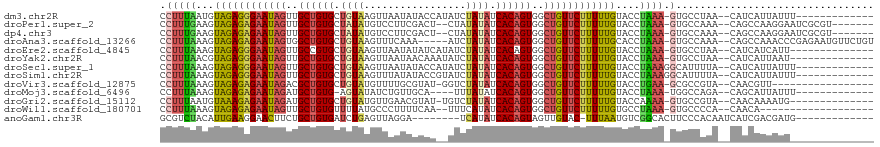
>dm3.chr2R 15548488 103 - 21146708 CCUUUAAUGUAGAGGGAAUAGUUGCUGUGCUGUAAGUUAAUAUACCAUAUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-GUGCCUAA--CAUCAUUAUUU------------- ....(((((.((((((((((((..(((((.((((..(...........)..)))).)))))..))))))))))))((((.....-))))....--...)))))...------------- ( -28.50, z-score = -3.04, R) >droPer1.super_2 129713 107 + 9036312 CCUUUGAAGUAGAGAGAAUAGUUGCUGUGCUAUAUGUCCUUCGACU--CUAUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-GUGCCAAA--CAGCCAAGGAAUCGCGU------- ((((.(..((((((((((((((..(((((.((((((((....))).--..))))).)))))..))))))))))))((((.....-))))...)--)..).))))........------- ( -34.80, z-score = -3.88, R) >dp4.chr3 12677072 107 - 19779522 CCUUUGAAGUAGAGAGAAUAGUUGCUGUGCUAUAUGUCCUUCGACU--CUAUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-GUGCCAAA--CAGCCAAGGAAUCGCGU------- ((((.(..((((((((((((((..(((((.((((((((....))).--..))))).)))))..))))))))))))((((.....-))))...)--)..).))))........------- ( -34.80, z-score = -3.88, R) >droAna3.scaffold_13266 10152497 111 + 19884421 CCUUUAAAGUAGAGAGAAUAGUGGCUGUGCUGUAAGUUUCAAA-----AUCUAUAUCACAGUGGCUGUUCUUUUUGCACCUAAA-GUGCCAAA--CAGCCAAACCCGAGAAUGUUCUGU .(((((..((((((((((((((.((((((.((((..((....)-----)..)))).)))))).))))))))))))))...))))-).((..((--((..(........)..))))..)) ( -33.30, z-score = -3.51, R) >droEre2.scaffold_4845 9748740 102 - 22589142 CCUUUAAAGUAGAGGGAAUAGUUGCCGUGCUGUAAGUUAAUAUAUCAUAUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-GUGCCUAA--CAUCAUCAUU-------------- .(((((...(((((((((((((..(.(((.((((..(...........)..)))).))).)..)))))))))))))....))))-).......--..........-------------- ( -22.80, z-score = -1.50, R) >droYak2.chr2R 7514917 102 + 21139217 CCUUUAACGUAGAGGGAAUAGUUGCUGUGCUGUAAGUUAAUAACAAAUAUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-GUGCCUAA--CAUCAUUAAU-------------- .(((((...(((((((((((((..(((((.((((..(...........)..)))).)))))..)))))))))))))....))))-).......--..........-------------- ( -27.40, z-score = -2.94, R) >droSec1.super_1 13063219 104 - 14215200 CCUUUAAAGUAGAGGGAAUAGUUGCUGUGCUGUAAGUUAAUAUACCAUAUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAAGGCAUUUUA--CAUCAUUAUUU------------- ((((((...(((((((((((((..(((((.((((..(...........)..)))).)))))..)))))))))))))....)))))).......--...........------------- ( -30.20, z-score = -3.57, R) >droSim1.chr2R 14253924 104 - 19596830 CCUUUAAAGUAGAGGGAAUAGUUGCUGUGCUGUAAGUUUAUAUACCGUAUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAAGGCAUUUUA--CAUCAUUAUUU------------- ((((((...(((((((((((((..(((((.((((..(...........)..)))).)))))..)))))))))))))....)))))).......--...........------------- ( -30.20, z-score = -3.56, R) >droVir3.scaffold_12875 16498860 98 + 20611582 CCUUUAAAGUAGAGAGAAUAGACGCUGUGCUGUAUGUUUUGCGUAU-GGUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUGAA-GCGCCGUA--CAACGUU----------------- .(((((...((((((((((((.((((((((((((((.....)))))-)).......))))))).))))))))))))....))))-).......--.......----------------- ( -27.91, z-score = -1.74, R) >droMoj3.scaffold_6496 20936289 98 - 26866924 CCUUUAAAGUAGAGAGAAUAGAUGCUGUG-AGUAUAUCUGUUGCA----UUUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA-UGGCCAGA--CAGCAUUAUUU------------- ((((((...((((((((((((..((((((-(.((((..((...))----..)))))))))))..))))))))))))....))))-.)).....--...........------------- ( -24.10, z-score = -1.42, R) >droGri2.scaffold_15112 2108207 101 - 5172618 CCUUUAAUGUAAAGAGAAUAGAUGCUGUGCUGUAUGUUGAACGUAU-UGUCUAUAUCACAGUGGCUGUUCUUUUUGUACCAAAA-GUGCCGUA--CAACAAAAUG-------------- .......((((((((((((((..((((((..(((((.....)))))-.........))))))..)))))))))))((((.....-))))....--..))).....-------------- ( -22.90, z-score = -1.56, R) >droWil1.scaffold_180701 1651310 95 - 3904529 CCUUUAAAGUAGAGAGAAUAGUUGCUGUGUUUUAUGCCCUUUUCAA--UUUCAUAUCACAGUGGCUGUUCUUUUUGUGCCUAAA-GUGCCCCA--CAACA------------------- .(((((..((((((((((((((..(((((...((((..........--...)))).)))))..))))))))))))))...))))-).......--.....------------------- ( -28.02, z-score = -4.24, R) >anoGam1.chr3R 43697851 97 - 53272125 GCGUCUACAUUGAAGGAACUUCUGCUGUGAUCUGAGUUAGGA--------UCAUAUCACAGUAGUUGUAC-UUUAAUGUCGGCACUUCCCACAAUCAUCGACGAUG------------- .((((.(((((((((..((..((((((((((.(((.......--------))).))))))))))..)).)-)))))))).((......)).........))))...------------- ( -32.60, z-score = -3.61, R) >consensus CCUUUAAAGUAGAGAGAAUAGUUGCUGUGCUGUAAGUUAAUAGAAC__AUCUAUAUCACAGUGGCUGUUCUUUUUGUACCUAAA_GUGCCAAA__CAUCAUUAUU______________ .........((((((((((((..((((((.((((.................)))).))))))..))))))))))))........................................... (-21.60 = -21.42 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:29 2011