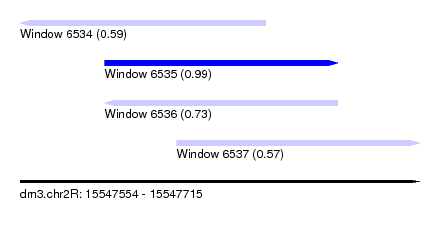
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,547,554 – 15,547,715 |
| Length | 161 |
| Max. P | 0.989794 |

| Location | 15,547,554 – 15,547,653 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.04 |
| Shannon entropy | 0.63909 |
| G+C content | 0.51490 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -12.24 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15547554 99 - 21146708 --GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGU---GGCCAGUUUG--CUGC---UGGCC-AAAUGCAAAACAAGCAAA --.........((.(((.......((((((.((--((((......)))))).))))))..-..((.(---(((((((...--..))---)))))-)...))...))).))... ( -39.70, z-score = -2.67, R) >droSim1.chr2R 14252978 99 - 19596830 --GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGU---GGCCAGUUUG--CUGC---UGGCC-AAAUGCAAAACAAGCAAA --.........((.(((.......((((((.((--((((......)))))).))))))..-..((.(---(((((((...--..))---)))))-)...))...))).))... ( -39.70, z-score = -2.67, R) >droSec1.super_1 13062223 99 - 14215200 --GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGU---GGCCAGUUUG--CUGC---UGGCC-AAAUGCAAAACAAGCAAA --.........((.(((.......((((((.((--((((......)))))).))))))..-..((.(---(((((((...--..))---)))))-)...))...))).))... ( -39.70, z-score = -2.67, R) >droYak2.chr2R 7513928 103 + 21139217 --GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCAUGGCCCACAGGCGU---GGCCAGUUUG--CUGCUGUUGGCC-AAAUGCAAAACAAGCAAA --.......((((.((((......(((((..((--((((......))))))..))))))))).))))---((((((....--......))))))-...(((.......))).. ( -37.60, z-score = -1.45, R) >droEre2.scaffold_4845 9747796 102 - 22589142 --GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCCACUCAUGGCCCG-AGGCGUGGCGGCCAGUUUG--CUGC---UGGCC-AAAUGCAAAACAAGCAAA --........(((((.(.(((..((((((....--.))))))....(((....))).)))-).)))))..(((((((...--..))---)))))-...(((.......))).. ( -37.80, z-score = -1.07, R) >droAna3.scaffold_13266 10151727 105 + 19884421 AAAUGCCACGCGCGUAUACCAACCGGUCAGAUGGCCUAGCCAGGACAGGAUUAGGAUGAGGUUGUG----GACUGCUU----UUGCUUUUGGCCAAAAUGCAAAACAAGCAAA .....(((((.((.(...((((((.(((...((((...)))).))).)).)).))...).))))))----)..(((((----((((((((....)))).))))...))))).. ( -26.90, z-score = 0.67, R) >dp4.chr3 12676406 91 - 19779522 -----------------ACGAAAU-GCCAAAUG--UGGUCCAUGACAAGGACAAGUCCGGACCGGGCUG-GACUGGACUGGACCACUGCUAGUC-CCAUGCAAAGGAAGUAAA -----------------.......-.((....(--(((((((.....((..((.(((((...)))))))-..))....))))))))(((.....-....)))..))....... ( -28.10, z-score = -0.90, R) >droPer1.super_2 129046 91 + 9036312 -----------------ACGAAAU-GCCAAAUG--UGGUCCAUGACAAGGACAAGUCCGGACCGGGCUG-GACUGGACUGGACCACUGCUAGUC-UCAUGCAAAGGAAGUAAA -----------------.......-.((....(--(((((((.....((..((.(((((...)))))))-..))....))))))))(((.....-....)))..))....... ( -28.10, z-score = -1.12, R) >consensus __GAAUUUAACGCGUGUGCGGAAUGGCCAAAAG__UUGGCCAAAACCAACUCAUGGCCCG_AGGCGU___GGCCAGUUUG__CUGC___UGGCC_AAAUGCAAAACAAGCAAA ................(((....(((((((.....))))))).............(((....)))((...(((((..............))))).....)).......))).. (-12.24 = -11.59 + -0.65)
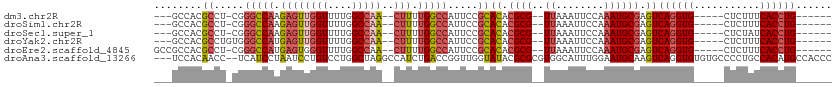
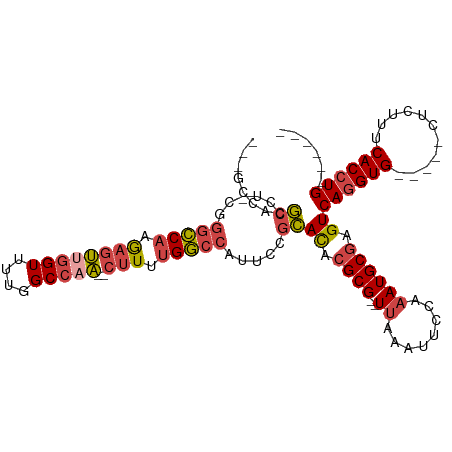
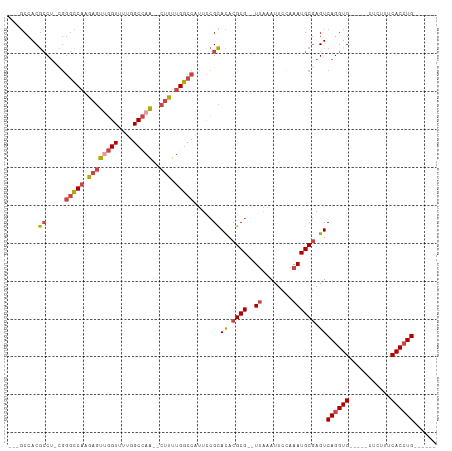
| Location | 15,547,588 – 15,547,682 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.35846 |
| G+C content | 0.55093 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.81 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15547588 94 + 21146708 ---GCCACGCCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGCACACGCG--UUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------ ---.....((..-..((((((((((((((....)))))--))))))))).....))((.((((--((........)))))).))((((((-----......))))))------ ( -39.70, z-score = -4.69, R) >droSim1.chr2R 14253012 94 + 19596830 ---GCCACGCCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGCACACGCG--UUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------ ---.....((..-..((((((((((((((....)))))--))))))))).....))((.((((--((........)))))).))((((((-----......))))))------ ( -39.70, z-score = -4.69, R) >droSec1.super_1 13062257 94 + 14215200 ---GCCACGCCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGCACACGCG--UUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUAUCACCUG------ ---.....((..-..((((((((((((((....)))))--))))))))).....))((.((((--((........)))))).))((((((-----......))))))------ ( -39.70, z-score = -4.78, R) >droYak2.chr2R 7513965 95 - 21139217 ---GCCACGCCUGUGGGCCAUGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGCACACGCG--UUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------ ---(((((....)))(((((.((((((((....)))))--))).))))).....))((.((((--((........)))))).))((((((-----......))))))------ ( -37.60, z-score = -3.49, R) >droEre2.scaffold_4845 9747830 97 + 22589142 GCCGCCACGCCU-CGGGCCAUGAGUGGGUUUUGGCCAA--CUUUUGGCCAUUCCGCACACGCG--UUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------ ...(((.((...-)))))..((.(((((...((((((.--....)))))).))))).))((((--((........))))))...((((((-----......))))))------ ( -33.90, z-score = -1.74, R) >droAna3.scaffold_13266 10151764 108 - 19884421 ---UCCACAACC--UCAUCCUAAUCCUGUCCUGGCUAGGCCAUCUGACCGGUUGGUAUACGCGCGUGGCAUUUGGAAUGCAAGUCAGGUGUGUGCCCCUGCCACAUGCCACCC ---.........--.................((((..((.(....).))(((.((..(((((((.((((..(((.....))))))).)))))))..)).)))....))))... ( -29.50, z-score = 1.01, R) >consensus ___GCCACGCCU_CGGGCCAAGAGUUGGUUUUGGCCAA__CUUUUGGCCAUUCCGCACACGCG__UUAAAUUCCAAAUGCGAGUCAGGUG_____CUCUUUCACCUG______ ...((...(((....)))........((...(((((((.....)))))))..))))((.((((..............)))).))((((((...........))))))...... (-20.19 = -19.81 + -0.38)
| Location | 15,547,588 – 15,547,682 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.35846 |
| G+C content | 0.55093 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15547588 94 - 21146708 ------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAA--CGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGUGGC--- ------((((((......-----)))))).(.(((.((((........--(((....)))....((((((.((--((((......)))))).))))))))-))))).)..--- ( -36.80, z-score = -2.63, R) >droSim1.chr2R 14253012 94 - 19596830 ------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAA--CGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGUGGC--- ------((((((......-----)))))).(.(((.((((........--(((....)))....((((((.((--((((......)))))).))))))))-))))).)..--- ( -36.80, z-score = -2.63, R) >droSec1.super_1 13062257 94 - 14215200 ------CAGGUGAUAGAG-----CACCUGACUCGCAUUUGGAAUUUAA--CGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGCCCG-AGGCGUGGC--- ------((((((......-----)))))).(.(((.((((........--(((....)))....((((((.((--((((......)))))).))))))))-))))).)..--- ( -36.80, z-score = -2.71, R) >droYak2.chr2R 7513965 95 + 21139217 ------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAA--CGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCAUGGCCCACAGGCGUGGC--- ------((((((......-----))))))..................(--(((.((((......(((((..((--((((......))))))..))))))))).))))...--- ( -36.40, z-score = -2.54, R) >droEre2.scaffold_4845 9747830 97 - 22589142 ------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAA--CGCGUGUGCGGAAUGGCCAAAAG--UUGGCCAAAACCCACUCAUGGCCCG-AGGCGUGGCGGC ------((((((......-----))))))..((((...(((.......--(((....)))...((((((....--.))))))....)))..((((.(...-.).)))))))). ( -32.10, z-score = -0.35, R) >droAna3.scaffold_13266 10151764 108 + 19884421 GGGUGGCAUGUGGCAGGGGCACACACCUGACUUGCAUUCCAAAUGCCACGCGCGUAUACCAACCGGUCAGAUGGCCUAGCCAGGACAGGAUUAGGAUGA--GGUUGUGGA--- (.(((((((.(((((((........)))).........))).))))))).).......((((((..(((.....(((((((......)).))))).)))--))))).)..--- ( -34.70, z-score = 0.68, R) >consensus ______CAGGUGAAAGAG_____CACCUGACUCGCAUUUGGAAUUUAA__CGCGUGUGCGGAAUGGCCAAAAG__UUGGCCAAAACCAACUCAUGGCCCG_AGGCGUGGC___ ......((((((...........))))))..((((..((((.........(((....)))...(((((((.....)))))))...))))......(((....))))))).... (-22.74 = -23.22 + 0.48)
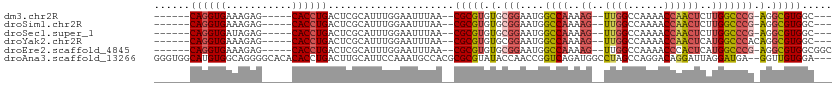
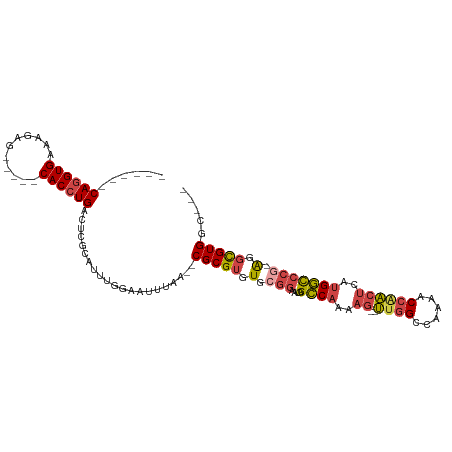
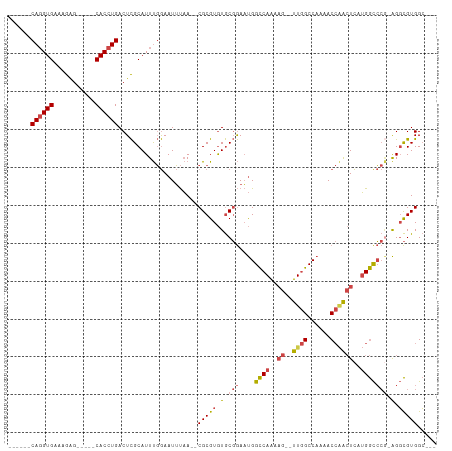
| Location | 15,547,617 – 15,547,715 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 98.37 |
| Shannon entropy | 0.02947 |
| G+C content | 0.52041 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15547617 98 + 21146708 GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((....((((((........))))))((((.(.(((((...(.(((....))))..))))).).)))).....))))... ( -32.70, z-score = -1.73, R) >droSim1.chr2R 14253041 98 + 19596830 GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((....((((((........))))))((((.(.(((((...(.(((....))))..))))).).)))).....))))... ( -32.70, z-score = -1.73, R) >droSec1.super_1 13062286 98 + 14215200 GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUAUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((....((((((........))))))((((.(.(((((...(.(((....))))..))))).).)))).....))))... ( -32.70, z-score = -1.81, R) >droYak2.chr2R 7513995 98 - 21139217 GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGAGUUGAGCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((.((.((((((........)))))).))).(((((((((((.....))))))))..(((((....)))))))))))... ( -31.30, z-score = -1.76, R) >droEre2.scaffold_4845 9747862 98 + 22589142 GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGCCCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((.((.((((((........)))))).))).(((((((((((.....)))))))).(((.....)))....))))))... ( -29.00, z-score = -0.62, R) >consensus GCCAACUUUUGGCCAUUCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAG ((((.....))))...((((.((.((((((........)))))).))).(((((((((((.....))))))))..(((((....)))))))))))... (-28.94 = -29.38 + 0.44)

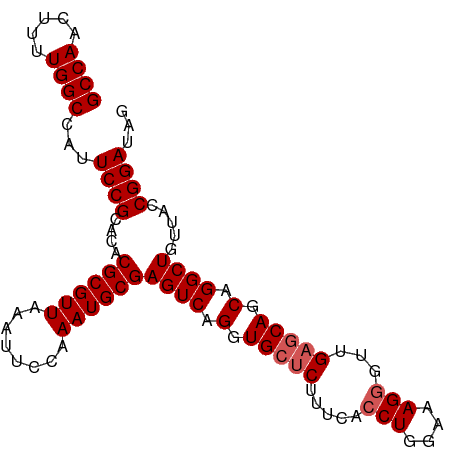
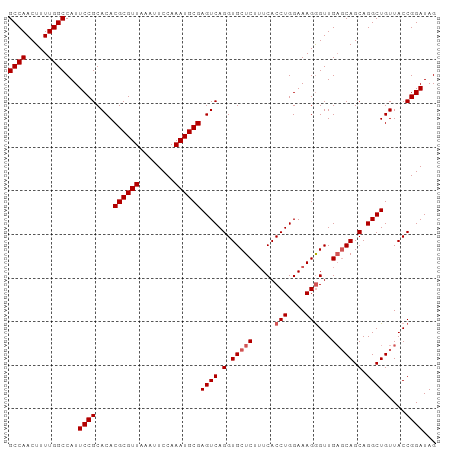
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:27 2011