
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,533,129 – 15,533,223 |
| Length | 94 |
| Max. P | 0.894505 |

| Location | 15,533,129 – 15,533,223 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.50233 |
| G+C content | 0.41619 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -9.59 |
| Energy contribution | -11.27 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
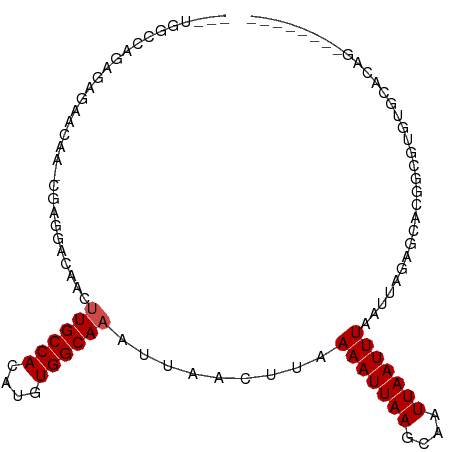
>dm3.chr2R 15533129 94 + 21146708 --CUGGCCAGAGAGGACAA-CGAGGAGAGGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGAGUGCACAG-------- --(((((((.(..((.(((-(........))))))...).)))).........(((((((((....))))))))).....((((.....)))).)))-------- ( -25.80, z-score = -3.37, R) >droSim1.chr2R 14236767 94 + 19596830 --CUGGCCAGAGAGGACAA-CGAGGAGAUGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGAGUGCACAG-------- --(((((((.(..((.(((-((......)))))))...).)))).........(((((((((....))))))))).....((((.....)))).)))-------- ( -25.60, z-score = -3.22, R) >droSec1.super_1 13047774 94 + 14215200 --CUGGCCAGAGAGGACAA-CGUGGAGAUGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGAGUGCACAG-------- --(((((((.(..((.(((-(((....))))))))...).)))).........(((((((((....))))))))).....((((.....)))).)))-------- ( -27.70, z-score = -3.40, R) >droYak2.chr2R 7499014 94 - 21139217 --GUGGACUGAGAGGACAA-CGAGGAGAAGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGAGUGCACAG-------- --((((.((....)).(((-(........)))))))).(((............(((((((((....))))))))).....((((.....))))))).-------- ( -22.60, z-score = -2.47, R) >droEre2.scaffold_4845 9731644 94 + 22589142 --CUGGCCAGAGAGGACAA-CGAGGAGAAGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGAGUGCACAG-------- --(((((((.(..((.(((-(........))))))...).)))).........(((((((((....))))))))).....((((.....)))).)))-------- ( -26.40, z-score = -3.74, R) >droAna3.scaffold_13266 10137062 91 - 19884421 -------------CUGGGC-GGAGGACAACUUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGUGUGCACAGCACAGCCC -------------....((-.(((...((.((((((....)))))).))..))).(((((((....))))))).......))..(((.(((((...)))))))). ( -25.10, z-score = -1.58, R) >droPer1.super_2 115539 86 - 9036312 ----------CUGCACUCA-CAAGGACAUCAUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAUAGCCGGGCAUGAGCACAG-------- ----------(((..((((-(..((.(....(((((....)))))..........(((((((....))))))).......)))..)..))))..)))-------- ( -18.30, z-score = -1.72, R) >dp4.chr3 12662886 86 + 19779522 ----------CUGCACUCA-CAAGGACAGCAUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAUAGCCGGGCAUGAGCACAG-------- ----------(((..((((-...(..(.((.(((((....)))))..........(((((((....))))))).......)).)..).))))..)))-------- ( -18.50, z-score = -1.29, R) >droGri2.scaffold_15112 2089542 97 + 5172618 AUGUGGCGGGCAACAAAGCCCGAGCGCAACUUGCCACAUGUGGCAAAUUAAGUUAAAAUUAAGCAAUUAAUUUAAUUAGUGCACUGUGUACAGGGGA-------- ......(((((......))))).((((...((((((....))))))....(((((((.((((....))))))))))).)))).(((....)))....-------- ( -30.60, z-score = -2.05, R) >droMoj3.scaffold_6496 20912793 94 + 26866924 --GGGGCAAACAAAACGAGGCGAACACAACUUGCCACAUGUGGCAAAUUAAGUUAAAAUUAAGCAAUUAAUUUAAUUAGUGCACUGCGUACAGGGA--------- --............(((((((.........((((((....))))))(((((....(((((((....)))))))..))))))).)).))).......--------- ( -18.30, z-score = -0.56, R) >droVir3.scaffold_12875 16477494 89 - 20611582 ---GGGCCAACAAAACGAG-----CACAACUUGCCACAUGUGGCAAAUUAAGUUAAAAUUAAGCAAUUAAUUUAAUUAGUGCACUGCGUACGGGGGA-------- ---...((..(...(((.(-----(((...((((((....))))))....(((((((.((((....))))))))))).))))....)))..)..)).-------- ( -22.60, z-score = -1.70, R) >consensus ___UGGCCAGAGAGAACAA_CGAGGACAACUUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAGAGCACGGCGUGUGCACAG________ ..............................((((((....)))))).........(((((((....)))))))................................ ( -9.59 = -11.27 + 1.68)

| Location | 15,533,129 – 15,533,223 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.50233 |
| G+C content | 0.41619 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -8.31 |
| Energy contribution | -8.49 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
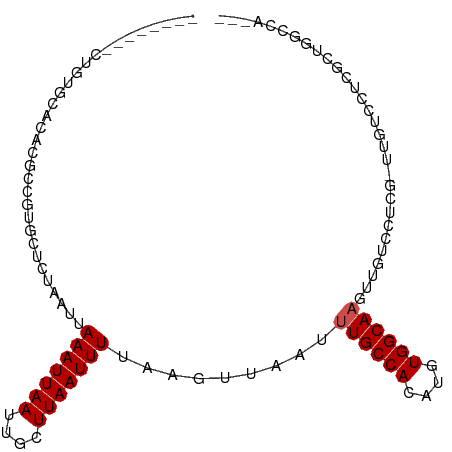
>dm3.chr2R 15533129 94 - 21146708 --------CUGUGCACUCGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACCUCUCCUCG-UUGUCCUCUCUGGCCAG-- --------..(.((((.....)))).)...(((((((((....))))))))).........((((...(.((((((........)-))).)).)..))))...-- ( -19.50, z-score = -2.55, R) >droSim1.chr2R 14236767 94 - 19596830 --------CUGUGCACUCGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACAUCUCCUCG-UUGUCCUCUCUGGCCAG-- --------..(.((((.....)))).)...(((((((((....))))))))).........((((...(.((((((........)-))).)).)..))))...-- ( -20.10, z-score = -2.51, R) >droSec1.super_1 13047774 94 - 14215200 --------CUGUGCACUCGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACAUCUCCACG-UUGUCCUCUCUGGCCAG-- --------..(.((((.....)))).)...(((((((((....))))))))).........((((...(.((((((........)-))).)).)..))))...-- ( -20.10, z-score = -2.07, R) >droYak2.chr2R 7499014 94 + 21139217 --------CUGUGCACUCGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACUUCUCCUCG-UUGUCCUCUCAGUCCAC-- --------(((.((((.....)))).....(((((((((....))))))))).......((((((....))))))..........-.........))).....-- ( -16.60, z-score = -2.10, R) >droEre2.scaffold_4845 9731644 94 - 22589142 --------CUGUGCACUCGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACUUCUCCUCG-UUGUCCUCUCUGGCCAG-- --------..(.((((.....)))).)...(((((((((....))))))))).........((((...(.((((((........)-))).)).)..))))...-- ( -20.00, z-score = -2.67, R) >droAna3.scaffold_13266 10137062 91 + 19884421 GGGCUGUGCUGUGCACACGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAAGUUGUCCUCC-GCCCAG------------- ((((.(..(.(((....))).)..)................(((((....)))))((((((((((....))))))))))......-))))..------------- ( -26.50, z-score = -2.09, R) >droPer1.super_2 115539 86 + 9036312 --------CUGUGCUCAUGCCCGGCUAUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAUGAUGUCCUUG-UGAGUGCAG---------- --------(((..((((((...(((.......(((((((((...........)))))))))((((....)))).....)))..))-))))..)))---------- ( -24.00, z-score = -2.46, R) >dp4.chr3 12662886 86 - 19779522 --------CUGUGCUCAUGCCCGGCUAUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAUGCUGUCCUUG-UGAGUGCAG---------- --------(((..((((((..((((.......(((((((((...........)))))))))((((....))))..))))..)..)-))))..)))---------- ( -26.90, z-score = -3.30, R) >droGri2.scaffold_15112 2089542 97 - 5172618 --------UCCCCUGUACACAGUGCACUAAUUAAAUUAAUUGCUUAAUUUUAACUUAAUUUGCCACAUGUGGCAAGUUGCGCUCGGGCUUUGUUGCCCGCCACAU --------.....(((....(((((.......(((((((....)))))))......(((((((((....))))))))))))))(((((......)))))..))). ( -27.80, z-score = -2.64, R) >droMoj3.scaffold_6496 20912793 94 - 26866924 ---------UCCCUGUACGCAGUGCACUAAUUAAAUUAAUUGCUUAAUUUUAACUUAAUUUGCCACAUGUGGCAAGUUGUGUUCGCCUCGUUUUGUUUGCCCC-- ---------.....(((.((((.((.......(((((((....))))))).(((.((((((((((....)))))))))).)))......)).)))).)))...-- ( -19.90, z-score = -1.89, R) >droVir3.scaffold_12875 16477494 89 + 20611582 --------UCCCCCGUACGCAGUGCACUAAUUAAAUUAAUUGCUUAAUUUUAACUUAAUUUGCCACAUGUGGCAAGUUGUG-----CUCGUUUUGUUGGCCC--- --------....(((.(((.((..(.......(((((((....)))))))......(((((((((....))))))))))..-----))))).....)))...--- ( -20.20, z-score = -1.42, R) >consensus ________CUGUGCACACGCCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAAGUUGUCCUCG_UUGUCCUCGCUGGCCA___ ................................(((((((....))))))).........((((((....)))))).............................. ( -8.31 = -8.49 + 0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:25 2011