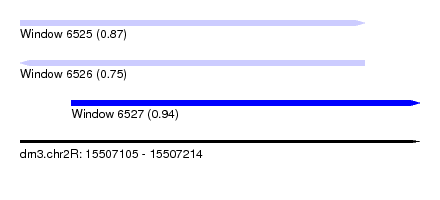
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,507,105 – 15,507,214 |
| Length | 109 |
| Max. P | 0.937916 |

| Location | 15,507,105 – 15,507,199 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.53540 |
| G+C content | 0.43720 |
| Mean single sequence MFE | -19.41 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.07 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
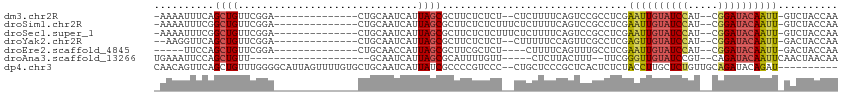
>dm3.chr2R 15507105 94 + 21146708 -AAAAUUUCAGCUGUUCGGA--------------CUGCAAUCAUUAGCGCUUCUCUCU--CUCUUUUCAGUCCGCCUCGAAUUGUAUCCAU--CGGAUACAAUU-GUCUACCAA -...............((((--------------(((.((..................--....)).))))))).....((((((((((..--.))))))))))-......... ( -18.75, z-score = -2.28, R) >droSim1.chr2R 14209728 96 + 19596830 -AAAAUUUCGGCUGUUCGGA--------------CUGCAAUCAUUAGCGCUUCUCUCUUUCUCUUUUCAGUCCGCCUCGAAUUGUAUCCAU--CGGAUACAAUU-GUCUACCAA -........(((....((((--------------(((.((.....((.(.....).))......)).))))))).....((((((((((..--.))))))))))-)))...... ( -21.00, z-score = -2.62, R) >droSec1.super_1 13021762 96 + 14215200 -AAAAUUUCGGCUGUUCGGA--------------CUGCAAUCAUUAGCGCUUCUCUCUUUCUCUUUUCAGUCCGCCUCGAAUUGUAUCCAU--CGGAUACAAUU-GUCUACCAA -........(((....((((--------------(((.((.....((.(.....).))......)).))))))).....((((((((((..--.))))))))))-)))...... ( -21.00, z-score = -2.62, R) >droYak2.chr2R 7470835 93 - 21139217 --AAGGUUCAGCUGUUCGGA--------------CUGCAAUCAUUAGCGCUUCUCUCU--CUUUUUCCAGUUCGCCUCGAGUUGUAUCCAU--CGGAUACAAUU-GACUACCAA --..(((((.......((((--------------(((.((..................--....)).))))))).....((((((((((..--.))))))))))-))..))).. ( -18.95, z-score = -0.63, R) >droEre2.scaffold_4845 9705160 88 + 22589142 -----UUCCAGCUGUUCGGA--------------CUGCAACCAUUAGCGCUUCGCUCU----CUUUUCAGUUUGCCUCGAAUUGUAUCCAU--CGGAUACAAUU-GACUACCAA -----...........((((--------------(((........((((...))))..----.....)))))))..((.((((((((((..--.))))))))))-))....... ( -18.56, z-score = -1.40, R) >droAna3.scaffold_13266 10110806 85 - 19884421 UGAAAUUCCAGCUGUU--------------------GCAAUCAUUAGCGCAUUUUGUU-----CUCUUACUUU--UUCGGGUUGUAUCCGU--CAGAUACAAUUCAACUAACAA .((((.....((((.(--------------------(....)).)))).......((.-----.....))..)--)))((((((((((...--..))))))))))......... ( -14.90, z-score = -1.18, R) >dp4.chr3 12633814 102 + 19779522 CAACAGUUCAGCUGUUUGGGGCAUUAGUUUUGUGCUGCAAUCAUUAUCGCCCCGUCCC--CUGCUCCCGCUCACUCUCUACCUUGCUCUGUUGCAGAUACAGAU---------- ((((((...(((.(..((((((..((((.(((.....)))..))))..))))))..).--..((....))..............)))))))))...........---------- ( -22.70, z-score = -0.08, R) >consensus _AAAAUUUCAGCUGUUCGGA______________CUGCAAUCAUUAGCGCUUCUCUCU__CUCUUUUCAGUCCGCCUCGAAUUGUAUCCAU__CGGAUACAAUU_GACUACCAA ..........((((..............................))))...............................((((((((((.....)))))))))).......... ( -9.94 = -10.07 + 0.13)
| Location | 15,507,105 – 15,507,199 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.53540 |
| G+C content | 0.43720 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15507105 94 - 21146708 UUGGUAGAC-AAUUGUAUCCG--AUGGAUACAAUUCGAGGCGGACUGAAAAGAG--AGAGAGAAGCGCUAAUGAUUGCAG--------------UCCGAACAGCUGAAAUUUU- (..((.(..-((((((((((.--..)))))))))).....(((((((.......--((.(.....).))........)))--------------))))..).))..)......- ( -23.46, z-score = -2.09, R) >droSim1.chr2R 14209728 96 - 19596830 UUGGUAGAC-AAUUGUAUCCG--AUGGAUACAAUUCGAGGCGGACUGAAAAGAGAAAGAGAGAAGCGCUAAUGAUUGCAG--------------UCCGAACAGCCGAAAUUUU- (((((.(..-((((((((((.--..)))))))))).....(((((((....((.....((.......)).....)).)))--------------))))..).)))))......- ( -25.30, z-score = -2.70, R) >droSec1.super_1 13021762 96 - 14215200 UUGGUAGAC-AAUUGUAUCCG--AUGGAUACAAUUCGAGGCGGACUGAAAAGAGAAAGAGAGAAGCGCUAAUGAUUGCAG--------------UCCGAACAGCCGAAAUUUU- (((((.(..-((((((((((.--..)))))))))).....(((((((....((.....((.......)).....)).)))--------------))))..).)))))......- ( -25.30, z-score = -2.70, R) >droYak2.chr2R 7470835 93 + 21139217 UUGGUAGUC-AAUUGUAUCCG--AUGGAUACAACUCGAGGCGAACUGGAAAAAG--AGAGAGAAGCGCUAAUGAUUGCAG--------------UCCGAACAGCUGAACCUU-- ..(((((((-(.((((((((.--..)))))))).....((((..((........--....))...))))..))))).(((--------------(.......)))).)))..-- ( -19.00, z-score = 0.07, R) >droEre2.scaffold_4845 9705160 88 - 22589142 UUGGUAGUC-AAUUGUAUCCG--AUGGAUACAAUUCGAGGCAAACUGAAAAG----AGAGCGAAGCGCUAAUGGUUGCAG--------------UCCGAACAGCUGGAA----- .......((-((((((((((.--..)))))))))).)).((((.((....))----..((((...)))).....))))..--------------((((......)))).----- ( -21.30, z-score = -0.59, R) >droAna3.scaffold_13266 10110806 85 + 19884421 UUGUUAGUUGAAUUGUAUCUG--ACGGAUACAACCCGAA--AAAGUAAGAG-----AACAAAAUGCGCUAAUGAUUGC--------------------AACAGCUGGAAUUUCA ...(((((((..(((((((..--...)))))))......--..........-----.......((((........)))--------------------).)))))))....... ( -14.90, z-score = -0.49, R) >dp4.chr3 12633814 102 - 19779522 ----------AUCUGUAUCUGCAACAGAGCAAGGUAGAGAGUGAGCGGGAGCAG--GGGACGGGGCGAUAAUGAUUGCAGCACAAAACUAAUGCCCCAAACAGCUGAACUGUUG ----------.(((.(((((((......)).))))).)))...(((((.(((.(--....)((((((.((....(((.....)))...)).)))))).....)))...))))). ( -29.70, z-score = -1.02, R) >consensus UUGGUAGAC_AAUUGUAUCCG__AUGGAUACAAUUCGAGGCGGACUGAAAAGAG__AGAGAGAAGCGCUAAUGAUUGCAG______________UCCGAACAGCUGAAAUUUU_ ........(.(((((((((((...))))))))))).).(((...((....))............(((........)))........................)))......... (-11.37 = -11.37 + 0.01)
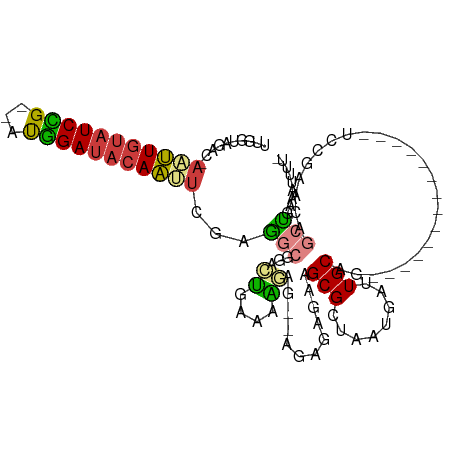
| Location | 15,507,119 – 15,507,214 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.30165 |
| G+C content | 0.46934 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -15.18 |
| Energy contribution | -14.90 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.937916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
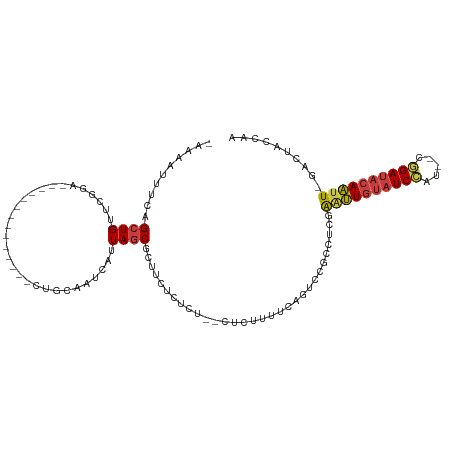
>dm3.chr2R 15507119 95 + 21146708 UCGGACUGCAAUCAUUAGCGCUUCUCUCU--CUCUUUUCAGUCCGCCUCGAAUUGUAUCCAUCGGAUACAAUU-GUCUACCAACUCUAACGGCGGCCG ..((((((.((..................--....)).))))))((((((((((((((((...))))))))))-((......)).....))).))).. ( -23.05, z-score = -2.62, R) >droSim1.chr2R 14209742 97 + 19596830 UCGGACUGCAAUCAUUAGCGCUUCUCUCUUUCUCUUUUCAGUCCGCCUCGAAUUGUAUCCAUCGGAUACAAUU-GUCUACCAACUCUAACGGCGGCAG ..((((((.((.....((.(.....).))......)).))))))((((((((((((((((...))))))))))-((......)).....))).))).. ( -24.10, z-score = -3.15, R) >droSec1.super_1 13021776 97 + 14215200 UCGGACUGCAAUCAUUAGCGCUUCUCUCUUUCUCUUUUCAGUCCGCCUCGAAUUGUAUCCAUCGGAUACAAUU-GUCUACCAACUCUAACGGCGGCAG ..((((((.((.....((.(.....).))......)).))))))((((((((((((((((...))))))))))-((......)).....))).))).. ( -24.10, z-score = -3.15, R) >droYak2.chr2R 7470848 95 - 21139217 UCGGACUGCAAUCAUUAGCGCUUCUCUCU--CUUUUUCCAGUUCGCCUCGAGUUGUAUCCAUCGGAUACAAUU-GACUACCAACUCUAACGGCGGCAG ..((((((.((..................--....)).))))))((((((((((((((((...))))))))))-((........))...))).))).. ( -21.05, z-score = -1.22, R) >droEre2.scaffold_4845 9705170 93 + 22589142 UCGGACUGCAACCAUUAGCGCUUCGCUCU----CUUUUCAGUUUGCCUCGAAUUGUAUCCAUCGGAUACAAUU-GACUACCAACUCUAACGGCGGCAG .....((((..((.((((.((...(((..----......)))..)).((.((((((((((...))))))))))-)).........)))).))..)))) ( -22.90, z-score = -2.08, R) >droAna3.scaffold_13266 10110821 84 - 19884421 ------UGCAAUCAUUAGCGCAUUUUGUU-----CUCUUACUUU--UUCGGGUUGUAUCCGUCAGAUACAAUUCAACUAACAACUA-ACCAGCGGCAG ------............(((.....((.-----.....))...--...((((((((((.....))))))))))............-....))).... ( -14.20, z-score = -1.20, R) >consensus UCGGACUGCAAUCAUUAGCGCUUCUCUCU__CUCUUUUCAGUCCGCCUCGAAUUGUAUCCAUCGGAUACAAUU_GACUACCAACUCUAACGGCGGCAG ..(((.(((........))).)))..................(((((...(((((((((.....))))))))).................)))))... (-15.18 = -14.90 + -0.28)
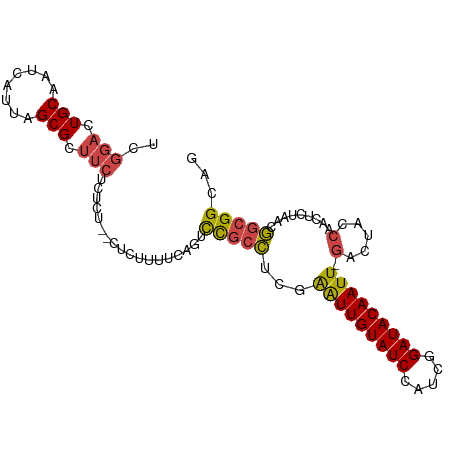
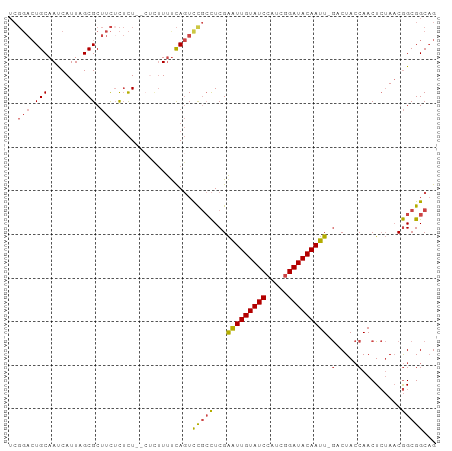
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:20 2011